Attenuation of Pseudomonas aeruginosa Quorum Sensing by Natural Products: Virtual Screening, Evaluation and Biomolecular Interactions
Abstract
:1. Introduction
2. Results
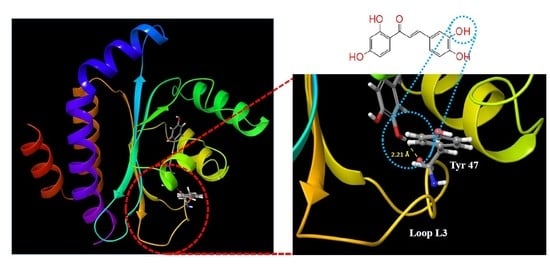
2.1. Molecular Docking Studies
2.2. In vitro studies
2.2.1. Influence on Biofilm Formation and Growth
2.2.2. Confocal and Scanning Electron Microscope Studies
2.3. Influence on Virulence Factors
2.4. Inhibition on QS-Related Gene Expression
2.5. Microscale Thermophoresis (MST) Analysis
2.6. Thermal Shift Assay (TSA)
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. In Silico Studies
4.3. In Vitro Assays
4.3.1. Biofilm Inhibition Assay
4.3.2. Pyocyanin Production
4.3.3. Elastase Activity
4.3.4. Rhamnolipid Assay
4.4. Confocal Laser Scanning Microscopy Analysis
4.5. SEM Analysis
4.6. qRT-PCR Studies
4.7. LasR-LBD Expression and Purification
4.8. Microscale Thermophoresis
4.9. Thermal Shift Assay (TSA)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| QS | Quorum sensing |
| QSIs | Quorum sensing inhibitors |
| C7X | Catechin-7-xyloside |
| MDR | Multi-drug resistant |
| AMR | Antimicrobial resistance |
| AIs | Autoinducers |
| AHL | Acyl homoserine lactone |
| LBD | Ligand binding domain |
| DBD | DNA-binding domain |
| 3OC12-HSL | N-(3-oxododecanoyl)-L-homoserine lactone |
| C4-HSL | N-butanoyl-L-homoserine lactone |
| CLSM | Confocal laser scanning microscopy |
| SEM | Scanning electron microscope |
| MST | Microscale thermophoresis |
| KD | Dissociation constant |
| TSA | Thermal shift assay |
| Tm B | Boltzmann Tm |
| ECR | Elastin congo red |
References
- Spaulding, C.N.; Klein, R.D.; Schreiber, H.L.; Janetka, J.W.; Hultgren, S.J. Precision antimicrobial therapeutics: The path of least resistance. NPJ Biofilms Microbiomes 2018, 27, 4. [Google Scholar] [CrossRef]
- Gupta, V.; Singhal, L. Antibiotics and Antimicrobial Resistance; Springer: Singapore, 2018; pp. 215–224. [Google Scholar]
- Durão, P.; Balbontín, R.; Gordo, I. Evolutionary mechanisms shaping the maintenance of antibiotic resistance. Trends Microbiol. 2018, 26, 677–691. [Google Scholar] [CrossRef] [Green Version]
- Padiyara, P.; Inoue, H.; Sprenger, M. Global governance mechanisms to address antimicrobial resistance. J. Infect. Dis. 2018, 11, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, A.S. A review of global initiatives to fight antibiotic resistance and recent antibiotics’ discovery. Acta Pharm. Sin. B 2016, 6, 552–556. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2018, 37, 177–192. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.; Song, H.; You, L. Signal discrimination by differential regulation of protein stability in quorum sensing. Am. J. Mol. Biol. 2008, 382, 1290–1297. [Google Scholar] [CrossRef] [Green Version]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [Green Version]
- O’Reilly, M.C.; Dong, S.H.; Rossi, F.M.; Karlen, K.M.; Kumar, R.S.; Nair, S.K.; Blackwell, H.E. Structural and biochemical studies of non-native agonists of the LasR quorum-sensing receptor reveal an L3 loop “out” conformation for LasR. Cell Chem. Biol. 2018, 25, 1128–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erickson, D.L.; Endersby, R.; Kirkham, A.; Stuber, K.; Vollman, D.D.; Rabin, H.R.; Mitchell, I.; Storey, D.G. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect. Immun. 2002, 70, 1783–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, P. Strategies for inhibiting quorum sensing. Emerg. Top. Life Sci. 2017, 1, 23–30. [Google Scholar] [CrossRef]
- Defoirdt, T. Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 2018, 26, 313–328. [Google Scholar] [CrossRef]
- Deryabin, D.; Galadzhieva, A.; Kosyan, D.; Duskaev, G. Plant-Derived Inhibitors of AHL-Mediated Quorum Sensing in Bacteria: Modes of Action. Int. J. Mol. Sci. 2019, 20, 5588. [Google Scholar] [CrossRef] [Green Version]
- Brackman, G.; Coenye, T. Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 2015, 21, 5–11. [Google Scholar] [CrossRef]
- Scoffone, V.C.; Trespidi, G.; Chiarelli, L.R.; Barbieri, G.; Buroni, S. Quorum Sensing as Antivirulence Target in Cystic Fibrosis Pathogens. Int. J. Mol. Sci. 2019, 20, 1838. [Google Scholar] [CrossRef] [Green Version]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [Green Version]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313. [Google Scholar] [CrossRef]
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Givskov, M. Quorum-sensing blockade as a strategy for enhancing host defences against bacterial pathogens. Philos. Trans. R. Soc. B 2007, 362, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- McCready, A.R.; Paczkowski, J.E.; Henke, B.R.; Bassler, B.L. Structural determinants driving homoserine lactone ligand selection in the Pseudomonas aeruginosa LasR quorum-sensing receptor. Proc. Natl. Acad. Sci. USA 2019, 116, 245–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paczkowski, J.E.; McCready, A.R.; Cong, J.P.; Li, Z.; Jeffrey, P.D.; Smith, C.D.; Henke, B.R.; Hughson, F.M.; Bassler, B.L. An autoinducer analog reveals an alternative mode of ligand binding for the LasR quorum-sensing receptor. ACS Chem. Biol. 2019, 14, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Ahumedo, M.; Díaz, A.; Vivas-Reyes, R. Theoretical and structural analysis of the active site of the transcriptional regulators LasR and TraR, using molecular docking methodology for identifying potential analogues of acyl homoserine lactones (AHLs) with anti-quorum sensing activity. Eur. J. Med. Chem. 2010, 45, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Paczkowski, J.E.; Mukherjee, S.; McCready, A.R.; Cong, J.P.; Aquino, C.J.; Kim, H.; Henke, B.R.; Smith, C.D.; Bassler, B.L. Flavonoids suppress Pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J. Biol. Chem. 2017, 292, 4064–4076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, J.; Sun, F.; Feng, W.; Sun, Y.; Qiu, X.; Xiong, L.; Liu, Y.; Chen, Y. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J. Appl. Microbiol. 2016, 120, 966–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudrappa, T.; Bais, H.P. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J. Agric. Food Chem. 2008, 56, 1955–1962. [Google Scholar] [CrossRef]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rasamiravaka, T.; Stevigny, C.; Duez, P.; Rajaonson, S.; Diallo, B.; Mol, A.; Baucher, M.; El Jaziri, M. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Microbiology 2011, 157, 2120–2132. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Lee, S.H.; Byun, Y.; Park, H.D. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci. Rep. 2015, 5, 8656. [Google Scholar] [CrossRef]
- Jayaraman, P.; Sakharkar, M.K.; Lim, C.S.; Tang, T.H.; Sakharkar, K.R. Activity and interactions of antibiotic and phytochemical combinations against Pseudomonas aeruginosa in vitro. Int. J. Biol. Sci. 2010, 6, 556. [Google Scholar] [CrossRef] [Green Version]
- Sarabhai, S.; Sharma, P.; Capalash, N. Ellagic acid derivatives from Terminalia chebula Retz. downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PLoS ONE 2013, 8, e53441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Packiavathy, I.A.S.V.; Agilandeswari, P.; Musthafa, K.S.; Pandian, S.K.; Ravi, A.V. Antibiofilm and quorum sensing inhibitory potential of Cuminum cyminum and its secondary metabolite methyl eugenol against Gram negative bacterial pathogens. Food Res. Int. 2012, 45, 85–92. [Google Scholar] [CrossRef]
- Kraft, L.; Serpell, L.C.; Atack, J.R. A biophysical approach to the identification of novel ApoE chemical probes. Biomolecules 2019, 9, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Y.; Nair, S.K. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chem. Biol. 2009, 16, 961–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinothkannan, R.; Zhong, L.; Wang, H.; Yu, G.; Zhang, Y.; Li, A. Virtual screening and biomolecular interactions of CviR-based quorum sensing inhibitors against Chromobacterium violaceum. Front. Cell. Infect. Microbiol. 2018, 8, 292. [Google Scholar] [CrossRef]
- He, J.B.; Lu, Q.; Cheng, Y.X. Two new sesquiterpenes from the resin of Toxicodendron vernicifluum. Helv. Chim. Acta 2015, 98, 1004–1008. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein− ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [Green Version]
- O’toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef]
- Karatuna, O.; Yagci, A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin. Microbiol. Infec. 2010, 16, 1770–1775. [Google Scholar] [CrossRef] [Green Version]
- Koch, A.K.; Kappeli, O.; Fiechter, A.; Reiser, J. Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants. J. Bacteriol. 1991, 173, 4212–4219. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T.; Liu, Y. N-acetylcysteine inhibit biofilms produced by Pseudomonas aeruginosa. BMC Microbiol. 2010, 10, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, V.K.; Mishra, A.; Jha, B. Anti-quorum sensing and anti-biofilm activity of Delftia tsuruhatensis extract by attenuating the quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2017, 7, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Li, Z.; Jia, R.; Hou, Y.; Yin, J.; Bian, X.; Li, A.; Müller, R.; Stewart, A.F.; Fu, J.; et al. RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat. Protoc. 2016, 11, 1175. [Google Scholar] [CrossRef] [PubMed]
- Lugo, A.C.; Daddaoua, A.; Ortega, A.; Morel, B.; Peña, A.I.; Espinosa-Urgel, M.; Krell, T. Purification and characterization of Pseudomonas aeruginosa LasR expressed in acyl-homoserine lactone free Escherichia coli cultures. Protein Expres. Purif. 2017, 130, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Yuan, Z.H.; Zhang, H.; Pan, Y.; Wu, Y.; Tian, X.Q.; Wang, F.F.; Wang, L.; Qian, W. Fatty acid DSF binds and allosterically activates histidine kinase RpfC of phytopathogenic bacterium Xanthomonas campestris pv. campestris to regulate quorum-sensing and virulence. PLoS Pathog. 2017, 13, e1006304. [Google Scholar] [CrossRef] [Green Version]












| Name | Structure | GScore | H Bonds | Bond Forming Amino Acids |
|---|---|---|---|---|
| 3OC12-HSL |  | −6.456 | 5 | Tyr 56 Trp 60 Asp 73 Ser 129 (2) |
| Catechin 7-xyloside (C7X) |  | −13.508 | 4 | Tyr 56 Tyr 64 Tyr 93 Leu 125 |
| Sappanol |  | −10.964 | 4 | Tyr 64 Thr 75 (2) Leu 125 |
| Butein |  | −9.245 | 3 | Tyr 47 Thr 75 Ser 129 |
| C30 |  | −4.244 | 1 | Ser 129 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, L.; Ravichandran, V.; Zhang, N.; Wang, H.; Bian, X.; Zhang, Y.; Li, A. Attenuation of Pseudomonas aeruginosa Quorum Sensing by Natural Products: Virtual Screening, Evaluation and Biomolecular Interactions. Int. J. Mol. Sci. 2020, 21, 2190. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21062190
Zhong L, Ravichandran V, Zhang N, Wang H, Bian X, Zhang Y, Li A. Attenuation of Pseudomonas aeruginosa Quorum Sensing by Natural Products: Virtual Screening, Evaluation and Biomolecular Interactions. International Journal of Molecular Sciences. 2020; 21(6):2190. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21062190
Chicago/Turabian StyleZhong, Lin, Vinothkannan Ravichandran, Na Zhang, Hailong Wang, Xiaoying Bian, Youming Zhang, and Aiying Li. 2020. "Attenuation of Pseudomonas aeruginosa Quorum Sensing by Natural Products: Virtual Screening, Evaluation and Biomolecular Interactions" International Journal of Molecular Sciences 21, no. 6: 2190. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21062190