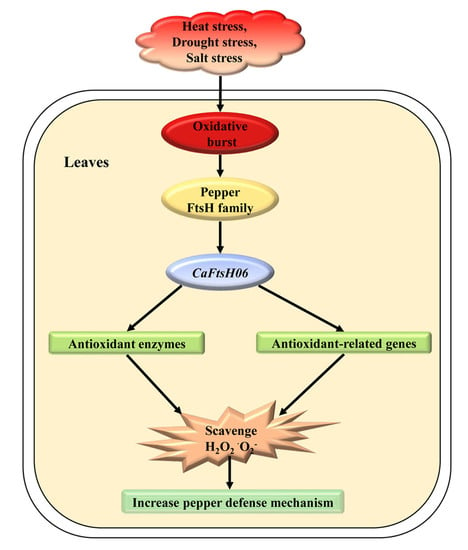
CaFtsH06, A Novel Filamentous Thermosensitive Protease Gene, Is Involved in Heat, Salt, and Drought Stress Tolerance of Pepper (Capsicum annuum L.)
Abstract
:1. Introduction
2. Results
2.1. The Expression Pattern of CaFtsH Genes in Pepper Across Different Tissues during Development and Stress Responses
2.2. Expression of CaFtsH06 under Heat Stress in Pepper
2.3. Structure Analysis of CaFtsH06 Proteins
2.4. Subcellular Localization of CaFtsH06 Proteins
2.5. Knockdown of CaFtsH06 Decreased Abiotic Stress Tolerance in Pepper
2.6. Ectopic Expression of CaFtsH06 Enhances the Abiotic Stress Tolerance in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Transcriptome Data Analysis of Pepper
4.3. Sequence Analysis and Identification of Conserved Domains
4.4. Subcellular Localization
4.5. Silencing of CaFtsH06 in Pepper
4.6. Construction of Transgenic Arabidopsis Line Expressing CaFtsH06
4.7. Plant Abiotic Stress Treatments
4.8. Biochemical Indices
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ROS | reactive oxygen species |
| PDS | phytoene desaturase |
| SOD | superoxide dismutase |
| NBT | nitro-blue tetrazolium |
| DAB | diaminobenzidine |
| H2O2 | hydrogen peroxide |
| ·O2− | superoxide |
| MDA | malondialdehyde |
| mg/g | milligram per liter |
| OE | overexpression |
| OE#7 | No. 7 Arabidopsis line with expressing CaFtsH06 |
| OE#9 | No. 9 Arabidopsis line with expressing CaFtsH06 |
| UBI | ubiquitin-conjugating protein |
| ORF | open reading frame |
| TRV | tobacco rattle virus |
| VIGS | virus-induced gene silencing |
References
- Zhai, Y.; Wang, H.; Liang, M.; Lu, M. Both silencing-and over-expression of pepper CaATG8c gene compromise plant tolerance to heat and salt stress. Environ. Exp. Bot. 2017, 141, 10–18. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Li, R.J.; Sun, J.T.; Ma, F.; Zhang, H.X.; Jin, J. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 2018, 8, 1–21. [Google Scholar]
- Allakhverdiev, S.I.; Kreslavski, V.D.; Klimov, V.V.; Los, D.A.; Carpentier, R.; Mohanty, P. Heat stress: An overview of molecular responses in photosynthesis. Photosynth. Res. 2008, 98, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; de Vos, R.C.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef]
- Van Breusegem, F.; Dat, J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2006, 141, 384–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [Green Version]
- Haq, S.U.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.X.; Zhang, H.X.; Wei, A.M.; Gong, Z.H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef] [Green Version]
- Gottesman, S.; Wickner, S.; Maurizi, M.R. Protein quality control: Triage by chaperones and proteases. Genes Dev. 1997, 11, 815–823. [Google Scholar] [CrossRef] [Green Version]
- Wagner, R.; Aigner, H.; Funk, C. FtsH proteases located in the plant chloroplast. Physiol. Plant 2012, 145, 203–214. [Google Scholar] [CrossRef]
- Ito, K.; Akiyama, Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 2005, 59, 211–231. [Google Scholar] [CrossRef]
- Schumann, W. FtsH—A single-chain charonin? FEMS Microbiol. Rev. 1999, 23, 1–11. [Google Scholar] [CrossRef]
- Tülek, A.; Özdemir, F.; Ramadhan, S. Cloning, expression and characterization of membrane bound FtsH protease of Geobacillus kaustophilus. Appl. Biochem. Microbiol. 2020, 56, 678–684. [Google Scholar] [CrossRef]
- Suzuki, C.K.; Rep, M.; van Dijl, J.M.; Suda, K.; Grivell, L.A.; Schatz, G. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem. Sci. 1997, 22, 118–123. [Google Scholar] [CrossRef]
- Janska, H.; Kwasniak, M.; Szczepanowska, J. Protein quality control in organelles—AAA/FtsH story. Biochim. Biophys. Acta BBA Mol. Cell Res. 2013, 1833, 381–387. [Google Scholar] [CrossRef] [Green Version]
- Adam, Z.; Adamska, I.; Nakabayashi, K.; Ostersetzer, O.; Haussuhl, K.; Manuell, A.; Zheng, B.; Vallon, O.; Rodermel, S.R.; Shinozaki, K. Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 2001, 125, 1912–1918. [Google Scholar] [CrossRef] [Green Version]
- Urantowka, A.; Knorpp, C.; Olczak, T.; Kolodziejczak, M.; Janska, H. Plant mitochondria contain at least two i-AAA-like complexes. Plant Mol. Biol. 2005, 59, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Herman, C.; Lecat, S.; D’Ari, R.; Bouloc, P. Regulation of the heat-shock response depends on divalent metal ions in an hfIB mutant of Escherichia coli. Mol. Microbiol. 1995, 18, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Okamoto, M.; Iwai, T.; Iwano, M.; Fukui, K.; Isogai, A.; Nakajima, N.; Ohashi, Y. Reduced levels of chloroplast FtsH protein in tobacco mosaic virus-infected tobacco leaves accelerate the hypersensitive reaction. Plant Cell 2000, 12, 917–932. [Google Scholar]
- Lindahl, M.; Spetea, C.; Hundal, T.; Oppenheim, A.B.; Adam, Z.; Andersson, B. The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 2000, 12, 419–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, W.; Zaltsman, A.; Adam, Z.; Takahashi, Y. Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 2003, 15, 2843–2855. [Google Scholar] [CrossRef] [Green Version]
- Ivashuta, S.; Imai, R.; Uchiyama, K.; Gau, M.; Shimamoto, Y. Changes in chloroplast FtsH-like gene during cold acclimation in alfalfa (Medicago sativa). J. Plant Physiol. 2002, 159, 85–90. [Google Scholar] [CrossRef]
- Żelisko, A.; García-Lorenzo, M.; Jackowski, G.; Jansson, S.; Funk, C. AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc. Natl. Acad. Sci. USA 2005, 102, 13699–13704. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka-Nishimura, M.; Yamamoto, Y. Quality control of photosystem II: The molecular basis for the action of FtsH protease and the dynamics of the thylakoid membranes. J. Photochem. Photobiol. B Biol. 2014, 137, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, M.; Uchida, S.; Mori, H.; Komayama, K.; Ohira, S.; Morita, N.; Nakanishi, T.; Yamamoto, Y. Quality control of photosystem II: Cleavage of reaction center D1 protein in spinach thylakoids by FtsH protease under moderate heat stress. J. Biol. Chem. 2006, 281, 21660–21669. [Google Scholar] [CrossRef] [Green Version]
- Okuno, T.; Ogura, T. FtsH protease-mediated regulation of various cellular functions. Regul. Proteolysis Microorg. 2013, 53–69. [Google Scholar] [CrossRef]
- Deuerling, E.; Paeslack, B.; Schumann, W. The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift. J. Bacteriol. 1995, 177, 4105–4112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deuerling, E.; Mogk, A.; Richter, C.; Purucker, M.; Schumann, W. The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol. Microbiol. 1997, 23, 921–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourdineaud, J.P.; Nehmé, B.; Tesse, S.; Lonvaud-Funel, A. The ftsH gene of the wine bacterium Oenococcus oeni is involved in protection against environmental stress. Appl. Environ. Microbiol. 2003, 69, 2512–2520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, A.Q.; Yi, S.Y.; Yang, J.Y.; Zhao, C.M.; Liu, J. Identification and characterization of a heat-inducible ftsH gene from tomato (Lycopersicon esculentum Mill.). Plant Sci. 2006, 170, 551–562. [Google Scholar] [CrossRef]
- Sedaghatmehr, M.; Mueller-Roeber, B.; Balazadeh, S. The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, E.; Fan, C.; Yang, Q.; Li, X.; Wan, B.; Dong, Y.; Wang, X.; Zhou, Y. Identification of heat responsive genes in Brassica napus siliques at the seed-filling stage through transcriptional profiling. PLoS ONE 2014, 9, e101914. [Google Scholar] [CrossRef]
- Xue, G.P.; Drenth, J.; McIntyre, C.L. TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 2015, 66, 1025–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, S.M.; Lim, F.L.; Finkler, A.; Fromm, H.; Slabas, A.R.; Knight, M.R. Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genom. 2014, 15, 456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.-T. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Zhai, Y.-F.; Gong, Z.H.; Lu, M.H. Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016, 252, 246–256. [Google Scholar] [CrossRef]
- Zhang, R.X.; Cheng, G.X.; Liu, G.T.; Chen, S.Y.; ul Haq, S.; Khan, A.; Li, Q.H.; Gong, Z.H. Assessing the functional role of color-related CaMYB gene under cold stress using virus-induced gene silencing in the fruit of pepper (Capsicum annuum L.). Sci. Hortic. 2020, 272, 109504. [Google Scholar] [CrossRef]
- Wang, H.; Niu, H.; Zhai, Y.; Lu, M. Characterization of BiP genes from pepper (Capsicum annuum L.) and the role of CaBiP1 in response to endoplasmic reticulum and multiple abiotic stresses. Front. Plant Sci. 2017, 8, 1122. [Google Scholar] [CrossRef] [Green Version]
- Tucker, P.A.; Sallai, L. The AAA+ superfamily—A myriad of motions. Curr. Opin. Struct. Biol. 2007, 17, 641–652. [Google Scholar] [CrossRef]
- Salehi-Lisar, S.Y.; Bakhshayeshan-Agdam, H. Drought stress in plants: Causes, consequences, and tolerance. In Drought Stress Tolerance in Plants; Springer: New York, NY, USA, 2016; Volume 1, pp. 1–16. [Google Scholar]
- Kato, Y.; Sakamoto, W. FtsH protease in the thylakoid membrane: Physiological functions and the regulation of protease activity. Front. Plant Sci. 2018, 9, 855. [Google Scholar] [CrossRef] [Green Version]
- Bujaldon, S.; Kodama, N.; Rappaport, F.; Subramanyam, R.; de Vitry, C.; Takahashi, Y.; Wollman, F.-A. Functional accumulation of antenna proteins in chlorophyll bless mutants of Chlamydomonas reinhardtii. Mol. Plant 2017, 10, 115–130. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Kim, C.; Xu, X.; Piskurewicz, U.; Dogra, V.; Singh, S.; Mahler, H.; Apel, K. Singlet oxygen-and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc. Natl. Acad. Sci. USA 2016, 113, E3792–E3800. [Google Scholar] [CrossRef] [Green Version]
- Dogra, V.; Duan, J.; Lee, K.P.; Lv, S.; Liu, R.; Kim, C. FtsH2-dependent proteolysis of EXECUTER1 is essential in mediating singlet oxygen-triggered retrograde signaling in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1145. [Google Scholar] [CrossRef] [Green Version]
- Janska, H.; Piechota, J.; Kwasniak, M. ATP-dependent proteases in biogenesis and maintenance of plant mitochondria. Biochim. Biophys. Acta BBA Bioenerg. 2010, 1797, 1071–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- War, A.R.; Paulraj, M.G.; War, M.Y.; Ignacimuthu, S. Differential defensive response of groundnut germplasms to Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae). J. Plant Interact. 2012, 7, 45–55. [Google Scholar] [CrossRef]
- Dai, H.; Wei, S.; Twardowska, I.; Han, R.; Xu, L. Hyperaccumulating potential of Bidens pilosa L. for Cd and elucidation of its translocation behavior based on cell membrane permeability. Environ. Sci. Pollut. Res. 2017, 24, 23161–23167. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.K.; Raikwar, S.; Tuteja, R.; Tuteja, N. Ectopic expression of phloem motor protein pea forisome PsSEO-F1 enhances salinity stress tolerance in tobacco. Plant Cell Rep. 2016, 35, 1021–1041. [Google Scholar] [CrossRef]
- Li, M.; Ji, L.; Jia, Z.; Yang, X.; Meng, Q.; Guo, S. Constitutive expression of CaHSP22. 5 enhances chilling tolerance in transgenic tobacco by promoting the activity of antioxidative enzymes. Funct. Plant Biol. 2018, 45, 575–585. [Google Scholar] [CrossRef]
- Burke, J.J.; Chen, J. Enhancement of reproductive heat tolerance in plants. PLoS ONE 2015, 10, e0122933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Khungar, L.; Shimphrui, R.; Tiwari, L.D.; Tripathi, G.; Sarkar, N.K.; Agarwal, S.K.; Agarwal, M.; Grover, A. AtHsp101 research sets course of action for the genetic improvement of crops against heat stress. J. Plant Biochem. Biotechnol. 2020, 29, 715–732. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Do Choi, Y.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruthvi, V.; Narasimhan, R.; Nataraja, K.N. Simultaneous expression of abiotic stress responsive transcription factors, AtDREB2A, AtHB7 and AtABF3 improves salinity and drought tolerance in peanut (Arachis hypogaea L.). PLoS ONE 2014, 9, e111152. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, S.; Bashir, K.; Matsui, A.; Tanaka, M.; Seki, M. Transcriptomic analysis of soil-grown Arabidopsis thaliana roots and shoots in response to a drought stress. Front. Plant Sci. 2016, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Seki, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration-and high-salinity-responsive gene expression. Plant Mol. Biol. 2000, 42, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.H.; Zhang, H.X.; Ali, M.; Gai, W.X.; Cheng, G.X.; Yu, Q.H.; Yang, S.B.; Li, X.X.; Gong, Z.H. A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper (Capsicum annuum L.). Plant Physiol. Biochem. 2019, 142, 151–162. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer Science and Business Media: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- de Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Nicholas, K.B.; Nicholas, H.B., Jr.; Deerfield, D.W. GeneDoc: Analysis and visualization of genetic variation. EMBNEW News 1997, 4, 14. [Google Scholar]
- Guo, M.; Zhai, Y.F.; Lu, J.P.; Chai, L.; Chai, W.G.; Gong, Z.H.; Lu, M.H. Characterization of CaHsp70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L. Int. J. Mol. Sci. 2014, 15, 19741–19759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Labra, M.; Vannini, C.; Grassi, F.; Bracale, M.; Balsemin, M.; Basso, B.; Sala, F. Genomic stability in Arabidopsis thaliana transgenic plants obtained by floral dip. Appl. Genet. 2004, 109, 1512–1518. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.J.; Cheng, G.X.; Khan, A.; Wei, A.M.; Yu, Q.-H.; Yang, S.B.; Luo, D.X.; Gong, Z.H. CaHSP16. 4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance. Protoplasma 2019, 256, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Yuan, W.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Zhao, J.; Liu, S.; et al. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 2011, 416, 24–30. [Google Scholar] [CrossRef]
- Stewart, R.R.; Bewley, J.D. Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol. 1980, 65, 245–248. [Google Scholar] [CrossRef] [Green Version]
- Arkus, K.A.; Cahoon, E.B.; Jez, J.M. Mechanistic analysis of wheat chlorophyllase. Arch. Biochem. Biophys. 2005, 438, 146–155. [Google Scholar] [CrossRef]
- Naser, L.; Kourosh, V.; Bahman, K.; Reza, A. Soluble sugars and proline accumulation play a role as effective indices for drought tolerance screening in Persian walnut (Juglans regia L.) during germination. Fruits 2010, 65, 97–112. [Google Scholar] [CrossRef] [Green Version]
- Sekulska-Nalewajko, J.; Goclawski, J.; Chojak-Kozniewska, J.; Kuzniak, E. Automated image analysis for quantification of reactive oxygen species in plant leaves. Methods 2016, 109, 114–122. [Google Scholar] [CrossRef] [PubMed]













Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, J.-J.; Zhang, R.-X.; Khan, A.; ul Haq, S.; Gai, W.-X.; Gong, Z.-H. CaFtsH06, A Novel Filamentous Thermosensitive Protease Gene, Is Involved in Heat, Salt, and Drought Stress Tolerance of Pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2021, 22, 6953. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22136953
Xiao J-J, Zhang R-X, Khan A, ul Haq S, Gai W-X, Gong Z-H. CaFtsH06, A Novel Filamentous Thermosensitive Protease Gene, Is Involved in Heat, Salt, and Drought Stress Tolerance of Pepper (Capsicum annuum L.). International Journal of Molecular Sciences. 2021; 22(13):6953. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22136953
Chicago/Turabian StyleXiao, Jing-Jing, Rui-Xing Zhang, Abid Khan, Saeed ul Haq, Wen-Xian Gai, and Zhen-Hui Gong. 2021. "CaFtsH06, A Novel Filamentous Thermosensitive Protease Gene, Is Involved in Heat, Salt, and Drought Stress Tolerance of Pepper (Capsicum annuum L.)" International Journal of Molecular Sciences 22, no. 13: 6953. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22136953