Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L.
Abstract
:1. Introduction
2. Results
2.1. Identification of PIN Family Members in T. aestivum
2.2. Chromosomal Distribution of TaPIN Genes
2.3. Gene Structure Analysis of TaPIN Genes
2.4. Conserved Motif Analysis of TaPIN Genes
2.5. Putative Cis-acting Regulatory Elements (CAREs) Analysis of TaPIN Genes
2.6. Gene Ontology (GO) Enrichment of TaPIN Genes
2.7. Expression Profiling of TaPIN Genes under Various Stress Conditions and Developmental Stages
2.8. Protein–Protein Network Analysis of the TaPIN Family Genes
3. Discussion
3.1. Identification and Evolution of TaPIN Gene Family in Wheat
3.2. Expression Profiling of TaPIN Genes in Various Tissues and Developmental Stages Indicating Their Role in Plant Growth and Development
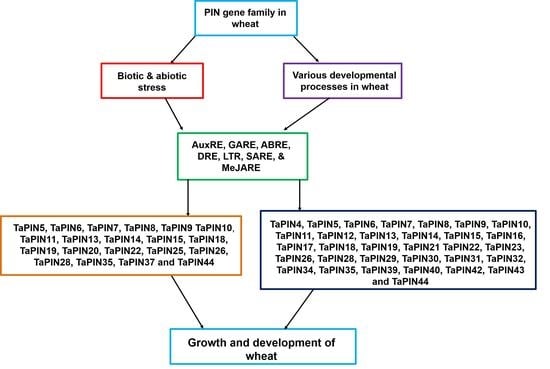
3.3. TaPIN Gene Family Members Respond to Biotic and Abiotic Stresses in Wheat
4. Materials and Methods
4.1. Identification of PIN Genes in the Wheat Genome
4.2. Chromosome Localization and Gene Duplication
4.3. Physico-Chemical Characteristics, Subcellular Localization, the Transmembrane Domain, and 3D Structure
4.4. Gene Structure, Gene Ontology, and Motif Analysis
4.5. Cis-acting Regulatory Elements (CAREs) Analysis and Protein Interaction Network
4.6. Expression Profiling of TaPIN Genes
4.7. Plant Material, Growth Conditions, Drought, and Heat Treatment
4.8. RNA Isolation and Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Benjamins, R.; Scheres, B. Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 2008, 59, 443–465. [Google Scholar] [CrossRef]
- Ganguly, A.; Park, M.; Kesawat, M.S.; Cho, H.-T. Functional Analysis of the Hydrophilic Loop in Intracellular Trafficking of Arabidopsis PIN-FORMED Proteins. Plant Cell 2014, 26, 1570–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y. Essential Roles of Local Auxin Biosynthesis in Plant Development and in Adaptation to Environmental Changes. Annu. Rev. Plant. Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.M.; Serre, N.B.; Oulehlová, D.; Vittal, P.; Fendrych, M. No Time for Transcription—Rapid Auxin Responses in Plants. Cold Spring Harb. Perspect. Biol. 2021, 13, a039891. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.J.; Marchant, A.; Green, H.G.; May, S.; Ward, S.P.; Millner, P.A.; Walker, A.; Schulz, B.; Feldmann, K.A.; Bennett, M.J.; et al. Arabidopsis AUX1 Gene: A Permease-Like Regulator of Root Gravitropism. Science 1996, 273, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Levernier, N.; Pouliquen, O.; Forterre, Y. An Integrative Model of Plant Gravitropism Linking Statoliths Position and Auxin Transport. Front. Plant. Sci. 2021, 12, 12. [Google Scholar] [CrossRef]
- Heisler, M.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of Auxin Transport and Gene Expression during Primordium Development Revealed by Live Imaging of the Arabidopsis Inflorescence Meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef] [Green Version]
- Scarpella, E.; Marcos, D.; Friml, J.; Berleth, T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006, 20, 1015–1027. [Google Scholar] [CrossRef] [Green Version]
- Leyser, O. Auxin Distribution and Plant Pattern Formation: How Many Angels Can Dance on the Point of PIN? Cell 2005, 121, 819–822. [Google Scholar] [CrossRef] [Green Version]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gra-dients establish the apical–basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Dubrovsky, J.G.; Sauer, M.; Napsucialy-Mendivil, S.; Ivanchenko, M.G.; Friml, J.; Shishkova, S.; Celenza, J.; Benkova, E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 2008, 105, 8790–8794. [Google Scholar] [CrossRef] [Green Version]
- Mattsson, J.; Ckurshumova, W.; Berleth, T. Auxin Signaling in Arabidopsis Leaf Vascular Development. Plant. Physiol. 2003, 131, 1327–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blakeslee, J.J.; Bandyopadhyay, A.; Peer, W.A.; Makam, S.N.; Murphy, A.S. Relocalization of the PIN1 Auxin Efflux Facilitator Plays a Role in Phototropic Responses. Plant. Physiol. 2004, 134, 28–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palme, K.; Dovzhenko, A.; Ditengou, F.A. Auxin transport and gravitational research: Perspectives. Protoplasma 2006, 229, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RE-SPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, B.B.; Stowe-Evans, E.L.; Harper, R.M.; Celaya, R.B.; Ljung, K.; Sandberg, G.; Liscum, E. Disruptions in AUX1-Dependent Auxin Influx Alter Hypocotyl Phototropism in Arabidopsis. Mol. Plant. 2008, 1, 129–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bainbridge, K.; Guyomarc’H, S.; Bayer, E.; Swarup, R.; Bennett, M.; Mandel, T.; Kuhlemeier, C. Auxin influx carriers stabilize phyllotactic patterning. Genes Dev. 2008, 22, 810–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarup, K.; Benková, E.; Swarup, R.; Casimiro, I.; Peret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef]
- Jones, A.; Kramer, E.M.; Knox, K.; Swarup, R.; Bennett, M.J.; Lazarus, C.M.; Leyser, H.M.O.; Grierson, C.S. Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 2008, 11, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Friml, J.; Benková, E.; Blilou, I.; Wisniewska, J.; Hamann, T.; Ljung, K.; Woody, S.; Sandberg, G.; Scheres, B.; Jürgens, G.; et al. AtPIN4 Mediates Sink-Driven Auxin Gradients and Root Patterning in Arabidopsis. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef] [Green Version]
- Dal Bosco, C.; Dovzhenko, A.; Palme, K. Intracellular auxin transport in pollen: PIN8, PIN5 and PILSPlant Signal. Behavior 2012, 7, 1504–1505. [Google Scholar]
- Zhou, J.-J.; Luo, J. The PIN-FORMED Auxin Efflux Carriers in Plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinhardt, D.; Mandel, T.; Kuhlemeier, C. Auxin regulates the initiation and radial position of plant lateral organs. Plant. Cell 2000, 12, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, Efflux-Dependent Auxin Gradients as a Common Module for Plant Organ Formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Petrasek, J.; Friml, J. Auxin transport routes in plant development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef] [Green Version]
- Konstantinova, N.; Korbei, B.; Luschnig, C. Auxin and Root Gravitropism: Addressing Basic Cellular Processes by Exploiting a Defined Growth Response. Int. J. Mol. Sci. 2021, 22, 2749. [Google Scholar] [CrossRef]
- Bartel, B. Auxin Biosynthesis. Annu. Rev. Plant. Biol. 1997, 48, 51–66. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin Biosynthesis and Its Role in Plant Development. Annu. Rev. Plant. Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [Green Version]
- Vanneste, S.; Friml, J. Auxin: A Trigger for Change in Plant Development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef]
- Swarup, R.; Péret, B. AUX/LAX family of auxin influx carriers—An overview. Front. Plant. Sci. 2012, 3, 225. [Google Scholar] [CrossRef] [Green Version]
- Vieten, A.; Vanneste, S.; Wiśniewska, J.; Benková, E.; Benjamins, R.; Beeckman, T.; Luschnig, C.; Friml, J. Functional redun-dancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 2005, 132, 4521–4531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geisler, M.; Murphy, A.S. The ABC of auxin transport: The role of p-glycoproteins in plant development. FEBS Lett. 2005, 580, 1094–1102. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Murphy, A.S. Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters inSchizosaccharomyces pombe. Plant. J. 2009, 59, 179–191. [Google Scholar] [CrossRef]
- Křeček, P.; Skůpa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zazimalova, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef] [Green Version]
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant. Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paponov, I.; Teale, W.D.; Trebar, M.; Blilou, I.; Palme, K. The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant. Sci. 2005, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Zažímalová, E.; Murphy, A.S.; Yang, H.; Hoyerová, K.; Hošek, P. Auxin transporters—why so many? Cold Spring Harb. Perspect. Biol. 2010, 2, a001552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdollahi Sisi, N.; Růžička, K. ER-Localized PIN Carriers: Regulators of Intracellular Auxin Homeostasis. Plants 2020, 9, 1527. [Google Scholar] [CrossRef] [PubMed]
- Friml, J. Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur. J. Cell Biol. 2010, 89, 231–235. [Google Scholar] [CrossRef]
- Mravec, J.; Skůpa, P.; Bailly, A.; Hoyerová, K.; Křeček, P.; Bielach, A.; Petrášek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.-D. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef]
- Ding, Z.; Wang, B.; Moreno, I.; Dupláková, N.; Simon, S.; Carraro, N.; Reemmer, J.; Pěnčík, A.; Chen, X.; Tejos, R.; et al. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012, 3, 941. [Google Scholar] [CrossRef] [Green Version]
- Simon, S.; Skůpa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klíma, P.; Čarná, M.; Rolčík, J.; De Rycke, R.; Moreno, I. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T. PIN proteins and the evolution of plant development. Trends Plant. Sci. 2015, 20, 498–507. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Zhao, P.; Wang, L.; Zhang, Y.; Wang, X.; Xiao, H.; Yu, J.; Xiao, G. The PIN gene family in cotton (Gossypium hirsutum): Genome-wide identification and gene expression analyses during root development and abiotic stress responses. BMC Genom. 2017, 18, 507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forestan, C.; Varotto, S. The Role of PIN Auxin Efflux Carriers in Polar Auxin Transport and Accumulation and Their Effect on Shaping Maize Development. Mol. Plant. 2012, 5, 787–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Yuan, Q.; Zhao, T.; Xiao, D.; Xu, X. Genome-wide identification and expression analysis of the PIN gene family during abiotic stress in tomato (Solanum lycopersium). Int. J. Agric. Biol. 2018, 20, 2481–2490. [Google Scholar]
- Wang, J.-R.; Hu, H.; Wang, G.-H.; Li, J.; Chen, J.-Y.; Wu, P. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol. Plant. 2009, 2, 823–831. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, J.; Wang, L.; Li, J.; Zheng, H.; Chen, J.; Lu, M. A survey of Populus PIN-FORMED family genes reveals their diversified expression patterns. J. Exp. Bot. 2014, 65, 2437–2448. [Google Scholar] [CrossRef]
- Wang, Y.; Chai, C.; Valliyodan, B.; Maupin, C.; Annen, B.; Nguyen, H.T. Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max). BMC Genom. 2015, 16, 951. [Google Scholar] [CrossRef] [Green Version]
- Pattison, R.; Catalá, C. Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant. J. 2012, 70, 585–598. [Google Scholar] [CrossRef]
- Xie, X.; Qin, G.; Si, P.; Luo, Z.; Gao, J.; Chen, X.; Zhang, J.; Wei, P.; Xia, Q.; Lin, F.; et al. Analysis of Nicotiana tabacum PIN genes identifies NtPIN4 as a key regulator of axillary bud growth. Physiol. Plant. 2017, 160, 222–239. [Google Scholar] [CrossRef]
- Shen, C.; Bai, Y.; Wang, S.; Zhang, S.; Wu, Y.; Chen, M.; Jiang, D.; Qi, Y. Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J. 2010, 277, 2954–2969. [Google Scholar] [CrossRef]
- Yang, C.; Wang, D.; Zhang, C.; Kong, N.; Ma, H.; Chen, Q. Comparative Analysis of the PIN Auxin Transporter Gene Family in Different Plant Species: A Focus on Structural and Expression Profiling of PINs in Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 3270. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 Family Gene, OsPIN1, involved in Auxin-dependent Adventitious Root Emergence and Tillering in Rice. Plant. Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef]
- Carraro, N.; Forestan, C.; Canova, S.; Traas, J.; Varotto, S. ZmPIN1a and ZmPIN1b Encode Two Novel Putative Candidates for Polar Auxin Transport and Plant Architecture Determination of Maize. Plant. Physiol. 2006, 142, 254–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Čovanová, M.; Sauer, M.; Rychtář, J.; Friml, J.; Petrášek, J.; Zažímalová, E. Overexpression of the auxin binding protein1 modulates PIN-dependent auxin transport in tobacco cells. PLoS ONE 2013, 8, e70050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Feng, F.; Liu, J.; Zhao, Q. Nitric Oxide Affects Rice Root Growth by Regulating Auxin Transport Under Nitrate Supply. Front. Plant. Sci. 2018, 9, 659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hartinger, C.; Wang, X.; Friml, J. Directional auxin fluxes in plants by intramolecular domain–domain coevolution of PIN auxin transporters. New Phytol. 2020, 227, 1406–1416. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Das, B.K.; Bhaganagare, G.R. Manorama Genome-wide identification, evolutionary and expression analyses of putative Fe–S biogenesis genes in rice (Oryza sativa). Genome 2012, 55, 571–583. [Google Scholar] [CrossRef]
- Gill, B.S.; Appels, R.; Botha, A.-M.; Buell, C.R.; Bennetzen, J.L.; Chalhoub, B.; Chumley, F.; Dvořák, J.; Iwanaga, M.; Keller, B.; et al. A Workshop Report on Wheat Genome Sequencing: International Genome Research on Wheat Consortium. Genetics 2004, 168, 1087–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Yadava, R.; Gollen, B.; Kumar, S.; Verma, R.K.; Yadav, S. Nutritional contents and medicinal properties of wheat: A review. Life Sci. Med. Res. 2011, 22, 1–10. [Google Scholar]
- Chen, Y.; Liu, T.; Tian, X.; Wang, X.; Li, M.; Wang, S.; Wang, Z. Effects of plastic film combined with straw mulch on grain yield and water use efficiency of winter wheat in Loess Plateau. Field Crop. Res. 2015, 172, 53–58. [Google Scholar] [CrossRef]
- Duan, Y.; Tao, X.; Zhao, H.; Xiao, X.; Li, M.; Wang, J.; Zhou, M. Activity of Demethylation Inhibitor Fungicide Metconazole on Chinese Fusarium graminearum Species Complex and Its Application in Carbendazim-Resistance Management of Fusarium Head Blight in Wheat. Plant. Dis. 2019, 103, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Afzal, F.; Chaudhari, S.K.; Gul, A.; Farooq, A.; Ali, H.; Nisar, S.; Sarfraz, B.; Shehzadi, K.J.; Mujeeb-Kazi, A. Bread Wheat (Triticum aestivum L.) Under Biotic and Abiotic Stresses: An Overview. In Crop Production and Global Environmental Issues; Hakeem, K., Ed.; Springer: Cham, Switzerland, 2015; pp. 293–317. [Google Scholar] [CrossRef]
- Kumar, R.; Masthigowda, M.H.; Kaur, A.; Bhusal, N.; Pandey, A.; Kumar, S.; Mishra, C.; Singh, G.P. Identification and characterization of multiple abiotic stress tolerance genes in wheat. Mol. Biol. Rep. 2020, 47, 8629–8643. [Google Scholar] [CrossRef] [PubMed]
- Fleury, D.; Jefferies, S.; Kuchel, H.; Langridge, P. Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 2010, 61, 3211–3222. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Nielsen, R. Mutation-Selection Models of Codon Substitution and Their Use to Estimate Selective Strengths on Codon Usage. Mol. Biol. Evol. 2008, 25, 568–579. [Google Scholar] [CrossRef] [Green Version]
- Sauer, M.; Kleine-Vehn, J. PIN-FORMED and PIN-LIKES auxin transport facilitators. Development 2019, 146, 15. [Google Scholar] [CrossRef] [Green Version]
- Sauer, M.; Grebe, M. Plant cell biology: PIN polarity maintained. Curr. Biol. 2021, 31, R449–R451. [Google Scholar] [CrossRef] [PubMed]
- Palovaara, J.; Hallberg, H.; Stasolla, C.; Luit, B.; Hakman, I. Expression of a gymnosperm PIN homologous gene correlates with auxin immunolocalization pattern at cotyledon formation and in demarcation of the procambium during Picea abies somatic embryo development and in seedling tissues. Tree Physiol. 2010, 30, 479–489. [Google Scholar] [CrossRef] [Green Version]
- Jordan, I.K.; Makarova, K.S.; Spouge, J.L.; Wolf, Y.I.; Koonin, E.V. Lineage-Specific Gene Expansions in Bacterial and Archaeal Genomes. Genome Res. 2001, 11, 555–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lespinet, O.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. The role of lineage-specific gene family expansion in the evolution of eu-karyotes. Genome Res. 2002, 12, 1048–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forestan, C.; Farinati, S.; Varotto, S. The maize PIN gene family of auxin transporters. Front. Plant. Sci. 2012, 3, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawton-Rauh, A. Evolutionary dynamics of duplicated genes in plants. Mol. Phylogenetics Evol. 2003, 29, 396–409. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [Green Version]
- Roumeliotis, E.; Kloosterman, B.; Oortwijn, M.; Visser, R.G.F.; Bachem, C.W.B. The PIN family of proteins in potato and their putative role in tuberization. Front. Plant. Sci. 2013, 4, 524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant. Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.L.; Sen, R.; Roeder, R.G. Enhancer–promoter communication and transcriptional regulation of Igh. Trends Immunol. 2011, 32, 532–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.L.; Singer, D.S. Core promoters in transcription: Old problem, new insights. Trends Biochem. Sci. 2015, 40, 165–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wong, C.-H.; Birnbaum, R.; Li, G.; Favaro, R.; Ngan, C.Y.; Lim, J.; Tai, E.; Poh, H.M.; Wong, E.; et al. Chromatin connectivity maps reveal dynamic promoter–enhancer long-range associations. Nat. Cell Biol. 2013, 504, 306–310. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Tang, Q.; Li, Y.; Zhang, Y.; Zhao, C.; Yan, J.; You, H. Folding/unfolding kinetics of G-quadruplexes upstream of the P1 promoter of the human BCL-2 oncogene. J. Biol. Chem. 2019, 294, 5890–5895. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Sahoo, D.K.; Dey, N.; Houtz, R.L.; Maiti, I.B. An intergenic region shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analyzed in transgenic Arabidopsis and to-bacco plants. PLoS ONE 2013, 8, e79622. [Google Scholar]
- Yue, R.; Tie, S.; Sun, T.; Zhang, L.; Yang, Y.; Qi, J.; Yan, S.; Han, X.; Wang, H.; Shen, C. Genome-Wide Identification and Expression Profiling Analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB Auxin Transporter Gene Families in Maize (Zea mays L.) under Various Abiotic Stresses. PLoS ONE 2015, 10, e0118751. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, A.; Lee, S.H.; Cho, M.; Lee, O.R.; Yoo, H.; Cho, H.-T. Differential Auxin-Transporting Activities of PIN-FORMED Proteins in Arabidopsis Root Hair Cells. Plant. Physiol. 2010, 153, 1046–1061. [Google Scholar] [CrossRef] [Green Version]
- Juárez, M.J.A.; Cárdenas, R.H.; Villa, J.N.S.; O’Connor, D.; Sluis, A.; Hake, S.; Ordaz-Ortiz, J.; Terry, L.; Simpson, J. Functionally different PIN proteins control auxin flux during bulbil development in Agave tequilana. J. Exp. Bot. 2015, 66, 3893–3905. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Jia, L.; Baluška, F.; Ding, G.; Shi, W.; Ye, N.; Zhang, J. PIN2 is required for the adaptation of Arabidopsis roots to alkaline stress by modulating proton secretion. J. Exp. Bot. 2012, 63, 6105–6114. [Google Scholar] [CrossRef] [Green Version]
- Shibasaki, K.; Uemura, M.; Tsurumi, S.; Rahman, A. Auxin Response in Arabidopsis under Cold Stress: Underlying Molecular Mechanisms. Plant. Cell 2010, 21, 3823–3838. [Google Scholar] [CrossRef] [Green Version]
- Kleine-Vehn, J.; Huang, F.; Naramoto, S.; Zhang, J.; Michniewicz, M.; Offringa, R.; Friml, J. PIN Auxin Efflux Carrier Polarity Is Regulated by PINOID Kinase-Mediated Recruitment into GNOM-Independent Trafficking in Arabidopsis. Plant. Cell 2010, 21, 3839–3849. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program. For Windows 95/98/NT, Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; pp. 95–98. [Google Scholar]
- Wang, Y.; Tang, H.; DeBarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kozlowski, L.P. IPC–isoelectric point calculator. Biol. Direct 2016, 11, 55. [Google Scholar] [CrossRef] [Green Version]
- Kozlowski, L.P. IPC 2.0: Prediction of isoelectric point and pKa dissociation constants. Nucleic Acids Res. 2021, 49, W285–W292. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2. 0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.K.; Kesawat, M.S.; Hong, C.B. One gene member of the ADP-ribosylation factor family is heat-inducible and enhances seed germination in Nicotiana tabacum. Genes Genom. 2017, 39, 1353–1365. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kim, D.K.; Zeba, N.; Suh, M.C.; Xia, X.; Hong, C.B. Ectopic RING zinc finger gene from hot pepper induces totally different genes in lettuce and tobacco. Mol. Breed. 2018, 38, 70. [Google Scholar] [CrossRef] [Green Version]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2(−ΔΔCT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]















| Proposed Gene Name | Gene ID | Genomic Location | Orientation | CDS Length (bp) | Intron Number | Protein Length (aa) | Molecular Weight (kDa) | Isoelectric Point (pI) | GRAVY | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|
| TaPIN1 | TraesCS1A02G415400 | 1A:574359017–574359996 | Reverse | 1800 | 2 | 599 | 63.57 | 8.632 | 0.354 | Plasma membrane |
| TaPIN2 | TraesCS1B02G445400 | 1B:665407466–665408862 | Reverse | 1794 | 2 | 597 | 63.37 | 8.989 | 0.297 | Plasma membrane |
| TaPIN3 | TraesCS1D02G422900 | 1D:478154308–478155466 | Reverse | 1791 | 2 | 596 | 63.18 | 9.12 | 0.321 | Plasma membrane |
| TaPIN4 | TraesCS3A02G231500 | 3A:432041807–432043538 | Forward | 1788 | 2 | 595 | 63.72 | 6.619 | 0.325 | Plasma membrane |
| TaPIN5 | TraesCS3A02G243700 | 3A:456920851–456922269 | Reverse | 1074 | 0 | 357 | 39.19 | 8.603 | 0.795 | Plasma membrane |
| TaPIN6 | TraesCS3A02G331300 | 3A:575870221–575871321 | Reverse | 1299 | 1 | 432 | 45.98 | 8.819 | 0.554 | Plasma membrane |
| TaPIN7 | TraesCS3A02G426700 | 3A:669623930–669624178 | Reverse | 1134 | 0 | 377 | 40.55 | 9.161 | 0.699 | Plasma membrane |
| TaPIN8 | TraesCS3B02G260700 | 3B:418660347–418660613 | Forward | 1830 | 1 | 609 | 65.38 | 7.54 | 0.243 | Plasma membrane |
| TaPIN9 | TraesCS3B02G276500 | 3B:446634435–446635854 | Forward | 1074 | 0 | 357 | 39.16 | 8.603 | 0.809 | Plasma membrane |
| TaPIN10 | TraesCS3B02G361500 | 3B:572835666–572837001 | Reverse | 1287 | 1 | 428 | 45.74 | 9.064 | 0.537 | Plasma membrane |
| TaPIN11 | TraesCS3B02G462900 | 3B:705706664–705706909 | Reverse | 1107 | 1 | 368 | 39.7 | 8.998 | 0.675 | Plasma membrane |
| TaPIN12 | TraesCS3D02G221900 | 3D:302847280–302848826 | Reverse | 1773 | 0 | 590 | 62.69 | 6.465 | 0.319 | Plasma membrane |
| TaPIN13 | TraesCS3D02G247700 | 3D:346727914–346729331 | Forward | 1074 | 0 | 357 | 39.1 | 8.603 | 0.809 | Plasma membrane |
| TaPIN14 | TraesCS3D02G324800 | 3D:437732979–437734144 | Reverse | 1296 | 3 | 431 | 45.92 | 8.689 | 0.592 | Plasma membrane |
| TaPIN15 | TraesCS3D02G421600 | 3D:533875425–533875673 | Reverse | 1137 | 1 | 378 | 40.49 | 9.147 | 0.714 | Plasma membrane |
| TaPIN16 | TraesCS4A02G188100 | 4A:466939085–466940349 | Reverse | 1707 | 1 | 568 | 60.78 | 8.361 | 0.336 | Plasma membrane |
| TaPIN17 | TraesCS4B02G130100 | 4B:170928310–170929402 | Forward | 1701 | 1 | 566 | 60.55 | 8.374 | 0.328 | Plasma membrane |
| TaPIN18 | TraesCS4D02G125300 | 4D:109470858–109471950 | Forward | 1701 | 2 | 566 | 60.59 | 8.361 | 0.327 | Plasma membrane |
| TaPIN19 | TraesCS5A02G284500 | 5A:492633348–492636528 | Reverse | 1098 | 0 | 365 | 38.67 | 8.28 | 0.714 | Plasma membrane |
| TaPIN20 | TraesCS5A02G285700 | 5A:493780035–493782506 | Forward | 1164 | 0 | 387 | 41.36 | 8.86 | 0.559 | Plasma membrane |
| TaPIN21 | TraesCS5A02G286000 | 5A:493815389–493817262 | Reverse | 1074 | 0 | 357 | 37.92 | 8.948 | 0.606 | Plasma membrane |
| TaPIN22 | TraesCS5B02G283500 | 5B:469121420–469124421 | Reverse | 1107 | 0 | 368 | 38.84 | 8.28 | 0.699 | Plasma membrane |
| TaPIN23 | TraesCS5B02G283600 | 5B:469292031–469299978 | Reverse | 1095 | 1 | 364 | 38.96 | 7.519 | 0.621 | Plasma membrane |
| TaPIN24 | TraesCS5B02G284900 | 5B:470436673–470438746 | Forward | 1134 | 1 | 377 | 40.51 | 8.149 | 0.606 | Plasma membrane |
| TaPIN25 | TraesCS5B02G285000 | 5B:470512790–470515294 | Forward | 1164 | 1 | 387 | 41.47 | 9.017 | 0.517 | Plasma membrane |
| TaPIN26 | TraesCS5D02G291800 | 5D:389457955–389460969 | Reverse | 1104 | 2 | 367 | 38.87 | 8.868 | 0.672 | Plasma membrane |
| TaPIN27 | TraesCS5D02G293000 | 5D:390372139–390374171 | Forward | 1134 | 1 | 377 | 40.4 | 8.147 | 0.621 | Plasma membrane |
| TaPIN28 | TraesCS5D02G293100 | 5D:390418535–390421740 | Forward | 1164 | 1 | 387 | 41.48 | 8.838 | 0.537 | Plasma membrane |
| TaPIN29 | TraesCS5D02G293300 | 5D:390504794–390506554 | Forward | 1134 | 1 | 377 | 40.03 | 9.059 | 0.634 | Plasma membrane |
| TaPIN30 | TraesCS6A02G308600 | 6A:543394829–543395365 | Forward | 1767 | 1 | 588 | 63.76 | 8.236 | 0.174 | Plasma membrane |
| TaPIN31 | TraesCS6B02G337300 | 6B:593711958–593712494 | Forward | 1566 | 3 | 521 | 56.79 | 7.407 | 0.011 | Plasma membrane |
| TaPIN32 | TraesCS6D02G287800 | 6D:397082369–397082905 | Forward | 1770 | 1 | 589 | 63.86 | 8.236 | 0.164 | Plasma membrane |
| TaPIN33 | TraesCS7A02G190600 | 7A:148415130–148415675 | Forward | 1764 | 1 | 587 | 64.02 | 8.652 | 0.176 | Plasma membrane |
| TaPIN34 | TraesCS7A02G258800 | 7A:250325040–250326673 | Reverse | 1104 | 1 | 367 | 39.23 | 8.126 | 0.654 | Plasma membrane |
| TaPIN35 | TraesCS7A02G492400 | 7A:681011797–681013386 | Reverse | 1899 | 1 | 632 | 67.3 | 9.256 | 0.254 | Plasma membrane |
| TaPIN36 | TraesCS7B02G095500 | 7B:109702420–109702965 | Forward | 1761 | 2 | 586 | 63.9 | 8.652 | 0.183 | Plasma membrane |
| TaPIN37 | TraesCS7B02G331500 | 7B:586716404–586721044 | Forward | 1221 | 2 | 406 | 43.87 | 6.619 | 0.536 | Plasma membrane |
| TaPIN38 | TraesCS7B02G359200 | 7B:621295197–621300495 | Forward | 1182 | 1 | 393 | 41.63 | 5.929 | 0.478 | Plasma membrane |
| TaPIN39 | TraesCS7B02G359300 | 7B:621396217–621403628 | Reverse | 1200 | 1 | 399 | 42.83 | 7.983 | 0.589 | Plasma membrane |
| TaPIN40 | TraesCS7B02G398100 | 7B:664108583–664110165 | Forward | 1899 | 1 | 632 | 67.26 | 9.256 | 0.252 | Plasma membrane |
| TaPIN41 | TraesCS7D02G191600 | 7D:146862944–146863489 | Forward | 1761 | 1 | 586 | 63.86 | 8.652 | 0.183 | Plasma membrane |
| TaPIN42 | TraesCS7D02G259700 | 7D:235396162–235396730 | Reverse | 1143 | 0 | 380 | 40.5 | 8.133 | 0.669 | Plasma membrane |
| TaPIN43 | TraesCS7D02G446900 | 7D:567514947–567515186 | Reverse | 858 | 0 | 285 | 30.22 | 6.456 | 0.496 | Plasma membrane |
| TaPIN44 | TraesCS7D02G478800 | 7D:589514264–589515825 | Forward | 1902 | 1 | 633 | 67.41 | 9.323 | 0.248 | Plasma membrane |
| Plant Species | Genome Size (Approx.) | Coding Genes | PIN Genes |
|---|---|---|---|
| Triticum aestivum (6n) | 17 Gb | 107,891 | 44 |
| Oryza sativa (2n) | 500 Mb | 37,960 | 12 |
| Arabidopsis thaliana (2n) | 135 MB | 27,655 | 8 |
| Zea mays (2n) | 2.4 Gb | 39,591 | 14 |
| Glycine max (2n) | 1.15 Gb | 55,897 | 23 |
| Nicotiana tabacum (4n) | 4.5 Gb | 61,526 | 29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, M.; Kherawat, B.S.; Dey, P.; Saha, D.; Singh, A.; Bhatia, S.K.; Ghodake, G.S.; Kadam, A.A.; Kim, H.-U.; Manorama; et al. Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021, 22, 7396. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147396
Kumar M, Kherawat BS, Dey P, Saha D, Singh A, Bhatia SK, Ghodake GS, Kadam AA, Kim H-U, Manorama, et al. Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L. International Journal of Molecular Sciences. 2021; 22(14):7396. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147396
Chicago/Turabian StyleKumar, Manu, Bhagwat Singh Kherawat, Prajjal Dey, Debanjana Saha, Anupama Singh, Shashi Kant Bhatia, Gajanan Sampatrao Ghodake, Avinash Ashok Kadam, Hyun-Uk Kim, Manorama, and et al. 2021. "Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L." International Journal of Molecular Sciences 22, no. 14: 7396. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147396