Free Radical-Mediated Protein Radical Formation in Differentiating Monocytes
Abstract
:1. Introduction
2. Results
2.1. Cell Differentiation by Phorbol 12-Myristate 13-Acetate
2.2. Cell Viability or Proliferation Assays
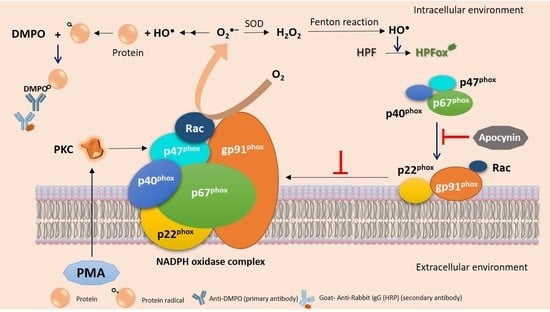
2.3. Hydroxyl Radical Imaging during Cell Differentiation
2.4. Detection of Hydroxyl Radical Formation by EPR Spin Trapping Spectroscopy
2.5. Protein-Centered Radicals Detection from U-937 Cells
2.6. Effect of Apocynin on Hydroxyl Radical Formation and NADPH Expression
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cell Culture and Treatment regime
4.3. Cell Proliferation Assay
4.4. Cell Membrane Integrity
4.5. Hydroxyl Radical Imaging Using Confocal Laser Scanning Microscopy (CLSM)
4.6. Hydroxyl Radical Detection Using EPR Spin Trapping Spectroscopy
4.7. Whole-Cell Protein Lysate Preparation
4.8. Western Blot Analysis of Nitrone Adducts
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Luan, Y.-Y.; Dong, N.; Xie, M.; Xiao, X.-Z.; Yao, Y.-M. The Significance and Regulatory Mechanisms of Innate Immune Cells in the Development of Sepsis. J. Interf. Cytokine Res. 2014, 34, 2–15. [Google Scholar] [CrossRef] [Green Version]
- Liggett, L.A.; Sankaran, V.G. Unraveling Hematopoiesis through the Lens of Genomics. Cell 2020, 182, 1384–1400. [Google Scholar] [CrossRef]
- Pennington, K.N.; Taylor, J.A.; Bren, G.D.; Paya, C.V. IκB Kinase-Dependent Chronic Activation of NF-κB Is Necessary for p21 WAF1/Cip1 Inhibition of Differentiation-Induced Apoptosis of Monocytes. Mol. Cell. Biol. 2001, 21, 1930–1941. [Google Scholar] [CrossRef] [Green Version]
- Pagliara, P.; Lanubile, R.; Dwikat, M.; Abbro, L.; Dini, L. Differentiation of monocytic U937 cells under static magnetic field exposure. Eur. J. Histochem. 2005, 49, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.; Sedlářová, M.; Balukova, A.; Ovsii, A.; Rác, M.; Křupka, M.; Kasai, S.; Pospíšil, P. Reactive Oxygen Species Imaging in U937 Cells. Front. Physiol. 2020, 11, 552569. [Google Scholar] [CrossRef]
- Prasad, A.; Kikuchi, H.; Inoue, K.Y.; Suzuki, M.; Sugiura, Y.; Sugai, T.; Tomonori, A.; Tada, M.; Kobayashi, M.; Matsue, T.; et al. Simultaneous Real-Time Monitoring of Oxygen Consumption and Hydrogen Peroxide Production in Cells Using Our Newly Developed Chip-Type Biosensor Device. Front. Physiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, E.M.; Park, Y.J.; Kang, H.S.; Cho, H.M.; Jun, D.Y.; Kim, Y.H. Expression of the apolipoprotein C-II gene during myelomonocytic differentiation of human leukemic cells. J. Leukoc. Biol. 2001, 69, 645–650. [Google Scholar] [PubMed]
- Yamamoto, T.; Sakaguchi, N.; Hachiya, M.; Nakayama, F.; Yamakawa, M.; Akashi, M. Role of catalase in monocytic differentiation of U937 cells by TPA: Hydrogen peroxide as a second messenger. Leukemia 2008, 23, 761–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamani, F.; Shahneh, F.Z.; Aghebati-Maleki, L.; Baradaran, B. Induction of CD14 Expression and Differentiation to Monocytes or Mature Macrophages in Promyelocytic Cell Lines: New Approach. Adv. Pharm. Bull. 2013, 3, 329–332. [Google Scholar] [CrossRef] [Green Version]
- Mendoza-Coronel, E.; Castañón-Arreola, M. Comparative evaluation ofin vitrohuman macrophage models for mycobacterial infection study. Pathog. Dis. 2016, 74, ftw052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lund, M.E.; To, J.; O’Brien, B.A.; Donnelly, S. The choice of phorbol 12-myristate 13-acetate differentiation protocol influences the response of THP-1 macrophages to a pro-inflammatory stimulus. J. Immunol. Methods 2016, 430, 64–70. [Google Scholar] [CrossRef]
- Kikuchi, H.; Prasad, A.; Matsuoka, R.; Aoyagi, S.; Matsue, T.; Kasai, S. Scanning Electrochemical Microscopy Imaging during Respiratory Burst in Human Cell. Front. Physiol. 2016, 7, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.M. Reactive oxygen species in phagocytic leukocytes. Histochem. Cell Biol. 2008, 130, 281–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pospíšil, P.; Prasad, A.; Rác, M. Role of reactive oxygen species in ultra-weak photon emission in biological systems. J. Photochem. Photobiol. B Biol. 2014, 139, 11–23. [Google Scholar] [CrossRef]
- Pospíšil, P.; Prasad, A.; Rác, M. Mechanism of the Formation of Electronically Excited Species by Oxidative Metabolic Processes: Role of Reactive Oxygen Species. Biomolecules 2019, 9, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutteridge, J.M.C.; Halliwell, B. Free Radicals and Antioxidants in the Year 2000: A Historical Look to the Future. Ann. N. Y. Acad. Sci. 2006, 899, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J. Free Radicals in Biology and Medicine, 4th ed.; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Radi, R. Oxygen radicals, nitric oxide, and peroxynitrite: Redox pathways in molecular medicine. Proc. Natl. Acad. Sci. USA 2018, 115, 5839–5848. [Google Scholar] [CrossRef] [Green Version]
- Panieri, E.; Santoro, M.M. ROS homeostasis and metabolism: A dangerous liason in cancer cells. Cell Death Dis. 2016, 7, e2253. [Google Scholar] [CrossRef]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, C.L.; Davies, M.J. Generation and propagation of radical reactions on proteins. Biochim. Biophys. Acta 2001, 1504, 196–219. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, C.L.; Davies, M.J. Detection, identification, and quantification of oxidative protein modifications. J. Biol. Chem. 2019, 294, 19683–19708. [Google Scholar] [CrossRef] [Green Version]
- Dean, R.T.; Fu, S.; Stocker, R.; Davies, M. Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J. 1997, 324, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berlett, B.S.; Stadtman, E.R. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997, 272, 20313–20316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, M.J. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 2003, 305, 761–770. [Google Scholar] [CrossRef]
- Di Mascio, P.; Martinez, G.R.; Miyamoto, S.; Ronsein, G.E.; Medeiros, M.H.G.; Cadet, J. Singlet Molecular Oxygen Reactions with Nucleic Acids, Lipids, and Proteins. Chem. Rev. 2019, 119, 2043–2086. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Prasad, A.; Sedlářová, M.; Pospíšil, P. Organic radical imaging in plants: Focus on protein radicals. Free Radic. Biol. Med. 2019, 130, 568–575. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, A.; Sedlářová, M.; Pospíšil, P. Characterization of Protein Radicals in Arabidopsis. Front. Physiol. 2019, 10, 958. [Google Scholar] [CrossRef] [Green Version]
- Mason, R.P. Using anti-5,5-dimethyl-1-pyrroline N-oxide (anti-DMPO) to detect protein radicals in time and space with immuno-spin trapping. Free Radic. Biol. Med. 2004, 36, 1214–1223. [Google Scholar] [CrossRef]
- Ramirez, D.C.; Mason, R.P. Immuno-Spin Trapping: Detection of Protein-Centered Radicals. Curr. Protoc. Toxicol. 2005, 24, 17.7.1–17.7.18. [Google Scholar] [CrossRef]
- Muñoz, M.D.; Gutierrez, L.J.; Delignat, S.; Russick, J.; Mejiba, S.E.G.; Lacroix-Desmazes, S.; Enriz, R.D.; Ramirez, D.C.; Gomez, S.E.; Enriz, D.R. The nitrone spin trap 5,5-dimethyl-1-pyrroline N-oxide binds to toll-like receptor-2-TIR-BB-loop domain and dampens downstream inflammatory signaling. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2019, 1865, 1152–1159. [Google Scholar] [CrossRef]
- Augustyniak, E.; Adam, A.; Wojdyla, K.; Rogowska-Wrzesinska, A.; Willetts, R.; Korkmaz, A.; Atalay, M.; Weber, D.; Grune, T.; Borsa, C.; et al. Validation of protein carbonyl measurement: A multi-centre study. Redox Biol. 2015, 4, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Verhoeckx, K.C.M.; Bijlsma, S.; de Groene, E.M.; Witkamp, R.F.; van der Greef, J.; Rodenburg, R.J.T. A combination of proteomics, principal component analysis and transcriptomics is a powerful tool for the identification of biomarkers for macrophage maturation in the U937 cell line. Proteomics 2004, 4, 1014–1028. [Google Scholar] [CrossRef]
- Traore, K.; Sharma, R.; Thimmulappa, R.K.; Watson, W.H.; Biswal, S.; Trush, M.A. Redox-regulation of Erk1/2-directed phosphatase by reactive oxygen species: Role in signaling TPA-induced growth arrest in ML-1 cells. J. Cell. Physiol. 2008, 216, 276–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefanska, J.; Pawliczak, R. Apocynin: Molecular Aptitudes. Mediat. Inflamm. 2008, 2008, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Hong, E.; Ding, B.; Jiang, W.; Zheng, S.; Xie, Z.; Tian, D.; Chen, Y. Inhibition of NOX4/ROS Suppresses Neuronal and Blood-Brain Barrier Injury by Attenuating Oxidative Stress After Intracerebral Hemorrhage. Front. Cell. Neurosci. 2020, 14. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, S.S.; Eligini, S.; Brambilla, M.; Tremoli, E.; Colli, S. Reactive oxygen species mediate cyclooxygenase-2 induction during monocyte to macrophage differentiation: Critical role of NADPH oxidase. Cardiovasc. Res. 2003, 60, 187–197. [Google Scholar] [CrossRef] [Green Version]
- Wardman, P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: Progress, pitfalls, and prospects. Free Radic. Biol. Med. 2007, 43, 995–1022. [Google Scholar] [CrossRef]
- Gomez-Mejiba, S.E.; Zhai, Z.; Akram, H.; Deterding, L.J.; Hensley, K.; Smith, N.; Towner, R.A.; Tomer, K.B.; Mason, R.P.; Ramirez, D.C. Immuno-spin trapping of protein and DNA radicals: “Tagging” free radicals to locate and understand the redox process. Free Radic. Biol. Med. 2009, 46, 853–865. [Google Scholar] [CrossRef] [Green Version]
- Mason, R.P. Imaging free radicals in organelles, cells, tissue, and in vivo with immuno-spin trapping. Redox Biol. 2016, 8, 422–429. [Google Scholar] [CrossRef] [Green Version]
- Kiningham, K.K.; Cardozo, Z.-A.; Cook, C.; Cole, M.P.; Stewart, J.C.; Tassone, M.; Coleman, M.C.; Spitz, D.R. All-trans-retinoic acid induces manganese superoxide dismutase in human neuroblastoma through NF-κB. Free Radic. Biol. Med. 2008, 44, 1610–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamiya, T.; Makino, J.; Hara, H.; Inagaki, N.; Adachi, T. Extracellular-superoxide dismutase expression during monocytic differentiation of U937 cells. J. Cell. Biochem. 2011, 112, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Ximenes, V.F.; Kanegae, M.P.; Rissato, S.R.; Galhiane, M.S. The oxidation of apocynin catalyzed by myeloperoxidase: Proposal for NADPH oxidase inhibition. Arch. Biochem. Biophys. 2007, 457, 134–141. [Google Scholar] [CrossRef]
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starr, T.; Bauler, T.; Malik-Kale, P.; Steele-Mortimer, O. The phorbol 12-myristate-13-acetate differentiation protocol is critical to the interaction of THP-1 macrophages with Salmonella Typhimurium. PLoS ONE 2018, 13, e0193601. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Nawaz, M.I.; Siddiquei, M.M.; Abu El-Asrar, A.M. Apocynin ameliorates NADPH oxidase 4 (NOX4) induced oxidative damage in the hypoxic human retinal Müller cells and diabetic rat retina. Mol. Cell. Biochem. 2021, 476, 2099–2109. [Google Scholar] [CrossRef]
- Castor, L.R.G.; Locatelli, K.A.; Ximenes, V.F. Pro-oxidant activity of apocynin radical. Free Radic. Biol. Med. 2010, 48, 1636–1643. [Google Scholar] [CrossRef]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasad, A.; Manoharan, R.R.; Sedlářová, M.; Pospíšil, P. Free Radical-Mediated Protein Radical Formation in Differentiating Monocytes. Int. J. Mol. Sci. 2021, 22, 9963. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189963
Prasad A, Manoharan RR, Sedlářová M, Pospíšil P. Free Radical-Mediated Protein Radical Formation in Differentiating Monocytes. International Journal of Molecular Sciences. 2021; 22(18):9963. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189963
Chicago/Turabian StylePrasad, Ankush, Renuka Ramalingam Manoharan, Michaela Sedlářová, and Pavel Pospíšil. 2021. "Free Radical-Mediated Protein Radical Formation in Differentiating Monocytes" International Journal of Molecular Sciences 22, no. 18: 9963. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189963