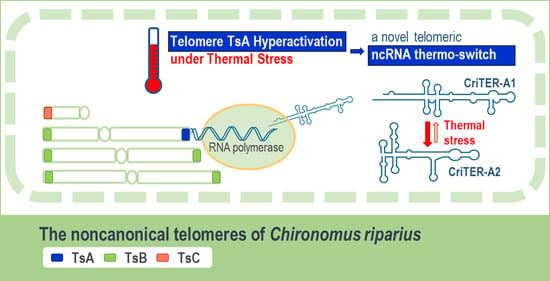
CriTER-A: A Novel Temperature-Dependent Noncoding RNA Switch in the Telomeric Transcriptome of Chironomus riparius
Abstract
:1. Introduction
2. Results and Discussion
2.1. RNA Structure Predictions: Remarkably Different Structures for the Two Telomeric Transcripts CriTER-A and CriTER-B
2.2. Telomeric RNAs of Chironomus riparius Adopt Two Alternate Conformations
2.3. CriTER-A and CriTER-B Fold into Different Secondary Structures
3. Conclusions
4. Materials and Methods
4.1. Sequences and Plasmids
4.2. DNA Templates and RNA Synthesis
4.3. Conformational Analysis by EMSA
4.4. SHAPE Analysis
4.5. Secondary Structure Prediction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef]
- Solovei, I.; Gaginskaya, E.R.; Macgregor, H.C. The arrangement and transcription of telomere DNA sequences at the ends of lampbrush chromosomes of birds. Chromosome Res. 1994, 2, 460–470. [Google Scholar] [CrossRef]
- Luke, B.; Panza, A.; Redon, S.; Iglesias, N.; Li, Z.; Lingner, J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 2008, 32, 465–477. [Google Scholar] [CrossRef]
- Vrbsky, J.; Akimcheva, S.; Watson, J.M.; Turner, T.L.; Daxinger, L.; Vyskot, B.; Aufsatz, W.; Riha, K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010, 6, e1000986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bah, A.; Wischnewski, H.; Shchepachev, V.; Azzalin, C.M. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2012, 40, 2995–3005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bah, A.; Azzalin, C.M. The telomeric transcriptome: From fission yeast to mammals. Int. J. Biochem. Cell Biol. 2012, 44, 1055–1059. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, J.; Cooper, J.P. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res. 2012, 40, 2956–2963. [Google Scholar] [CrossRef] [PubMed]
- Cusanelli, E.; Chartrand, P. Telomeric noncoding RNA: Telomeric repeat-containing RNA in telomere biology. Wiley Interdiscip. Rev. RNA 2014, 5, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Bettin, N.; Oss Pegorar, C.; Cusanelli, E. The emerging roles of TERRA in telomere maintenance and genome stability. Cells 2019, 8, 246. [Google Scholar] [CrossRef] [Green Version]
- Kordyukova, M.Y.; Kalmykova, A.I. Nature and functions of telomeric transcripts. Biochem. Mosc. 2019, 84, 137–146. [Google Scholar] [CrossRef]
- Mason, J.M.; Frydrychova, R.C.; Biessmann, H. Drosophila telomeres: An exception providing new insights. BioEssays 2008, 30, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Casacuberta, E. Drosophila: Retrotransposons making up telomeres. Viruses 2017, 9, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmona, M.J.; Morcillo, G.; Galler, R.; Martínez-Salas, E.; de la Campa, A.G.; Díez, J.L.; Edström, J.E. Cloning and molecular characterization of a telomeric sequence from a temperature-induced Balbiani ring. Chromosoma 1985, 92, 108–115. [Google Scholar] [CrossRef]
- López, C.C.; Nielsen, L.; Edström, J.E. Terminal long tandem repeats in chromosomes form Chironomus pallidivittatus. Mol. Cell. Biol. 1996, 16, 3285–3290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosén, M.; Edström, J.-E. DNA structures common for chironomid telomeres terminating with complex repeats. Insect Mol. Biol. 2000, 9, 341–347. [Google Scholar] [CrossRef]
- Rosén, M.; Castillejo-López, C.; Edström, J.-E. Telomere terminating with centromere-specific repeats is closely associated with a transposon derived gene in Chironomus pallidivittatus. Chromosoma 2002, 110, 532–541. [Google Scholar] [CrossRef]
- Rosén, M.; Edström, J.-E. Chromosome ends in Chironomus tentans do not have long single-stranded overhangs characterizing canonical telomeres. Chromosome Res. 2002, 10, 21–31. [Google Scholar] [CrossRef]
- Morcillo, G.; Santa-Cruz, M.C.; Díez, J.L. Temperature-induced Balbiani rings in Chironomus thummi. Chromosoma 1981, 83, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Barettino, D.; Morcillo, G.; Díez, J.-L. Induction of the heat-shock response by carbon dioxide in Chironomus thummi. Cell Differ. 1988, 23, 27–36. [Google Scholar] [CrossRef]
- Martínez-Guitarte, J.L.; Díez, J.L.; Morcillo, G. transcription and activation under environmental stress of the complex telomeric repeats of Chironomus thummi. Chromosome Res. 2008, 16, 1085–1096. [Google Scholar] [CrossRef]
- Martínez-Guitarte, J.-L.; Planelló, R.; Morcillo, G. Overexpression of long non-coding RNAs following exposure to xenobiotics in the aquatic midge Chironomus riparius. Aquat. Toxicol. 2012, 110–111, 84–90. [Google Scholar] [CrossRef]
- Martínez, J.L.; Edström, J.E.; Morcillo, G.; Diez, J.L. Telomeres in Chironomus thummi are characterized by different subfamilies of complex DNA repeats. Chromosoma 2001, 110, 221–227. [Google Scholar] [CrossRef]
- Martínez-Guitarte, J.L.; de la Fuente, M.; Morcillo, G. Telomeric transcriptome from Chironomus riparius (Diptera), a species with noncanonical telomeres. Insect Mol. Biol. 2014, 23, 367–380. [Google Scholar] [CrossRef]
- Martinez, J.L.; Sanchez-Elsner, T.; Morcillo, G.; Diez, J.L. Heat shock regulatory elements are present in telomeric repeats of Chironomus thummi. Nucleic Acids Res. 2001, 29, 4760–4766. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Komiyama, M. structure, function and targeting of human telomere RNA. Methods 2012, 57, 100–105. [Google Scholar] [CrossRef]
- Merino, E.J.; Wilkinson, K.A.; Coughlan, J.L.; Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005, 127, 4223–4231. [Google Scholar] [CrossRef] [PubMed]
- Gherghe, C.M.; Shajani, Z.; Wilkinson, K.A.; Varani, G.; Weeks, K.M. strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 2008, 130, 12244–12245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-López, C.; Barroso-delJesus, A.; Berzal-Herranz, A. The chaperone-like activity of the hepatitis C virus IRES and CRE elements regulates genome dimerization. Sci. Rep. 2017, 7, 43415. [Google Scholar] [CrossRef] [PubMed]
- Hajdin, C.E.; Bellaousov, S.; Huggins, W.; Leonard, C.W.; Mathews, D.H.; Weeks, K.M. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. USA 2013, 110, 5498–5503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-López, C.; Berzal-Herranz, A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell. Mol. Life Sci. 2012, 69, 103–113. [Google Scholar] [CrossRef]
- Romero-López, C.; Díaz-González, R.; Berzal-Herranz, A. Inhibition of hepatitis C virus internal ribosome entry site-mediated translation by an RNA targeting the conserved IIIf domain. Cell. Mol. Life Sci. 2007, 64, 2994–3006. [Google Scholar] [CrossRef] [Green Version]
- Romero-López, C.; Barroso-delJesus, A.; García-Sacristán, A.; Briones, C.; Berzal-Herranz, A. The folding of the hepatitis C virus internal ribosome entry site depends on the 3′-end of the viral genome. Nucleic Acids Res. 2012, 40, 11697. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Ruiz, M.; Sievers, D.; García-López, P.A.; Berzal-Herranz, A. The antisense sequence of the HIV-1 TAR stem-loop structure covalently linked to the hairpin ribozyme enhances its catalytic activity against two artificial substrates. Antisense Nucleic Acid Drug Dev. 1999, 9, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Barroso-Deljesus, A.; García-Sacristán, A.; Briones, C.; Berzal-Herranz, A. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3′x-tail region. Nucleic Acids Res. 2014, 42, 567–582. [Google Scholar] [CrossRef]
- Deigan, K.E.; Li, T.W.; Mathews, D.H.; Weeks, K.M. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA 2009, 106, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Steen, K.-A.; Rice, G.M.; Weeks, K.M. Fingerprinting noncanonical and tertiary rna structures by differential SHAPE reactivity. J. Am. Chem. Soc. 2012, 134, 13160–13163. [Google Scholar] [CrossRef] [Green Version]
- Rice, G.M.; Leonard, C.W.; Weeks, K.M. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA 2014, 20, 846–854. [Google Scholar] [CrossRef] [Green Version]
- Zuker, M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucl. Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Siederdissen, C.H.Z.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellaousov, S.; Reuter, J.S.; Seetin, M.G.; Mathews, D.H. RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013, 41, W471–W474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Title 1Oligonucleotide | Sequence (5′-3′) 1 |
| T7pCriTERA | TAATACGACTCACTATAGGGTTCCATAGGGGGGTACATGGGATA |
| T7pCriTERB | TAATACGACTCACTATAGGGTTCCATAGGGGGGTACAGGGGGTT |
| asCriTERAB | AATTCTAGAAAAATCGAGTTT |
| T7p_CriTERA_5cas | TAATACGACTCACTATAGGGACCAACCGGCGCGCCCACAGGACGTCAA GTTCCCGGGCCGTGGTCAGATTCCATAGGGGGGTACATGGGATA |
| T7p_CriTERB_5cas | TAATACGACTCACTATAGGGACCAACCGGCGCGCCCACAGGACGTCAA GTTCCCGGGCCGTGGTCAGATTCCATAGGGGGGTACAGGGGGTT |
| asCriTERAB_3cas | TTTTTCTTTGAGGTTTAGGATTCGTGCCAGTGGTGCACGGTCTACAATTCT AGAAAAATCGAGTTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero-López, C.; Berzal-Herranz, A.; Martínez-Guitarte, J.L.; de la Fuente, M. CriTER-A: A Novel Temperature-Dependent Noncoding RNA Switch in the Telomeric Transcriptome of Chironomus riparius. Int. J. Mol. Sci. 2021, 22, 10310. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms221910310
Romero-López C, Berzal-Herranz A, Martínez-Guitarte JL, de la Fuente M. CriTER-A: A Novel Temperature-Dependent Noncoding RNA Switch in the Telomeric Transcriptome of Chironomus riparius. International Journal of Molecular Sciences. 2021; 22(19):10310. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms221910310
Chicago/Turabian StyleRomero-López, Cristina, Alfredo Berzal-Herranz, José Luis Martínez-Guitarte, and Mercedes de la Fuente. 2021. "CriTER-A: A Novel Temperature-Dependent Noncoding RNA Switch in the Telomeric Transcriptome of Chironomus riparius" International Journal of Molecular Sciences 22, no. 19: 10310. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms221910310