SUMOylation Potentiates ZIC Protein Activity to Influence Murine Neural Crest Cell Specification
Abstract
:1. Introduction
2. Results
2.1. The ZIC Proteins Can Be SUMOylated at a Conserved Lysine in the ZF-NC Domain
2.2. The Zic Genes Cooperate during Murine Neural Crest Cell Specification
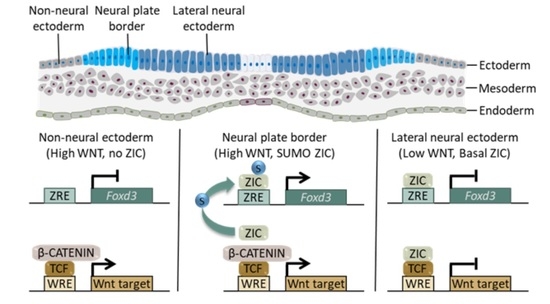
2.3. SUMOylation Promotes ZIC Trans-Activation of the Foxd3 Enhancer
2.4. The ZIC Proteins Are Context-Dependent Inhibitors of Canonical WNT-Dependent Transcription In Vitro and In Vivo
2.5. SUMOylation Decreases ZIC1, ZIC2, and ZIC3 Inhibition of TCF-Dependent Transcription
2.6. ZIC Protein SUMOylation Is Enhanced in a High-WNT Environment
3. Discussion
4. Materials and Methods
4.1. Mouse Strains and Husbandry
4.2. Embryo Collection and Pre-Processing
4.3. Whole Mount in Situ Hybridization
4.4. Whole Mount Immunofluorescence and Confocal Microscopy
4.5. Plasmids
4.6. Cell Culture, Subcellular Fractionation, SDS-PAGE, and Western Blotting
4.7. SUMOylation Assays
4.8. Luciferase Reporter Assays
4.9. Antibodies
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Le Douarin, N.M.; Dupin, E. The Neural Crest, a Fourth Germ Layer of the Vertebrate Embryo: Significance in Chordate Evolution. In Neural Crest Cells: Evolution, Development and Disease; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 3–26. ISBN 9780124017306. [Google Scholar]
- Simões-Costa, M.; Bronner, M.E. Establishing neural crest identity: A gene regulatory recipe. Development 2015, 142, 242–257. [Google Scholar] [CrossRef] [Green Version]
- Stuhlmiller, T.J.; García-Castro, M.I. Current perspectives of the signaling pathways directing neural crest induction. Cell. Mol. Life Sci. 2012, 69, 3715–3737. [Google Scholar] [CrossRef] [Green Version]
- Groves, A.K.; LaBonne, C. Setting appropriate boundaries: Fate, patterning and competence at the neural plate border. Dev. Biol. 2014, 389, 2–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barriga, E.H.; Trainor, P.A.; Bronner, M.; Mayor, R. Animal models for studying neural crest development: Is the mouse different? Development 2015, 142, 1555–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saint-Jeannet, J.P.; He, X.; Varmus, H.E.; Dawid, I.B. Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proc. Natl. Acad. Sci. USA 1997, 94, 13713–13718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Castro, M.I.; Marcelle, C.; Bronner-Fraser, M. Ectodermal Wnt function as a neural crest inducer. Science 2002, 297, 848–851. [Google Scholar] [CrossRef]
- Elkouby, Y.M.; Elias, S.; Casey, E.S.; Blythe, S.A.; Tsabar, N.; Klein, P.S.; Root, H.; Liu, K.J.; Frank, D. Mesodermal Wnt signaling organizes the neural plate via Meis3. Development 2010, 137, 1531–1541. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Zhang, R.; Lin, X.; Xu, X. Wnt3a regulates the development of cardiac neural crest cells by modulating expression of cysteine-rich intestinal protein 2 in rhombomere 6. Circ. Res. 2008, 102, 831–839. [Google Scholar] [CrossRef] [Green Version]
- LaBonne, C.; Bronner-Fraser, M. Neural crest induction in Xenopus: Evidence for a two signal model. Development 1998, 125, 2403–2414. [Google Scholar] [CrossRef]
- Steventon, B.; Araya, C.; Linker, C.; Kuriyama, S.; Mayor, R. Differential requirements of BMP and Wnt signalling during gastrulation and neurulation define two steps in neural crest induction. Development 2009, 136, 771–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, J.L.; Bonner, J.; Modrell, M.; Ragland, J.W.; Moon, R.T.; Dorsky, R.I.; Raible, D.W. Reiterated Wnt signaling during zebrafish neural crest development. Development 2004, 131, 1299–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Amerongen, R.; Berns, A. Knockout mouse models to study Wnt signal transduction. Trends Genet. 2006, 22, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Takada, S.; Stark, K.L.; Shea, M.J.; Vassileva, G.; McMahon, J.A.; McMahon, A.P. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994, 8, 174–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greco, T.L.; Takada, S.; Newhouse, M.M.; McMahon, J.A.; McMahon, A.P.; Camper, S.A. Analysis of the vestigial tail mutation demonstrates that Wnt-3a gene dosage regulates mouse axial development. Genes Dev. 1996, 10, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, Y.; Fujimori, T.; McMahon, A.P.; Takada, S. Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Dev. Biol. 1997, 183, 234–242. [Google Scholar] [CrossRef] [Green Version]
- Nakaya, M.A.; Biris, K.; Tsukiyama, T.; Jaime, S.; Rawls, J.A.; Yamaguchi, T.P. Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development 2005, 132, 5425–5436. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.M.K.; Tole, S.; Grove, E.; McMahon, A.P. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development 2000, 127, 457–467. [Google Scholar] [CrossRef]
- Carmona-Fontaine, C.; Acuña, G.; Ellwanger, K.; Niehrs, C.; Mayor, R. Neural crests are actively precluded from the anterior neural fold by a novel inhibitory mechanism dependent on Dickkopf1 secreted by the prechordal mesoderm. Dev. Biol. 2007, 309, 208–221. [Google Scholar] [CrossRef] [Green Version]
- Mašek, J.; Machoň, O.; Kořínek, V.; Taketo, M.M.; Kozmik, Z. Tcf7l1 protects the anterior neural fold from adopting the neural crest fate. Development 2016, 143, 2206–2216. [Google Scholar] [CrossRef] [Green Version]
- Luan, Z.; Liu, Y.; Stuhlmiller, T.J.; Marquez, J.; García-Castro, M.I. SUMOylation of Pax7 is essential for neural crest and muscle development. Cell. Mol. Life Sci. 2013, 70, 1793–1806. [Google Scholar] [CrossRef] [Green Version]
- Taylor, K.M.; LaBonne, C. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev. Cell 2005, 9, 593–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.A.J.; Wu, M.H.; Yan, C.H.; Chau, B.K.H.; So, H.; Ng, A.; Chan, A.; Cheah, K.S.E.; Briscoe, J.; Cheung, M. Phosphorylation of Sox9 is required for neural crest delamination and is regulated downstream of BMP and canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 2882–2887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, P.C.; Taylor-Jaffe, K.M.; Nordin, K.M.; Prasad, M.S.; Lander, R.M.; LaBonne, C. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J. Cell Biol. 2012, 198, 799–813. [Google Scholar] [CrossRef]
- Ali, R.G.; Bellchambers, H.M.; Warr, N.; Ahmed, J.N.; Barratt, K.S.; Neill, K.; Diamand, K.E.M.; Arkell, R.M. WNT responsive SUMOylation of ZIC5 promotes murine neural crest cell development via multiple effects on transcription. J. Cell Sci. 2021, 134, jcs.256792. [Google Scholar] [CrossRef]
- Milet, C.; Maczkowiak, F.; Roche, D.D.; Monsoro-Burq, A.H. Pax3 and Zic1 drive induction and differentiation of multipotent, migratory, and functional neural crest in Xenopus embryos. Proc. Natl. Acad. Sci. USA 2013, 110, 5528–5533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plouhinec, J.L.; Roche, D.D.; Pegoraro, C.; Figueiredo, A.L.; Maczkowiak, F.; Brunet, L.J.; Milet, C.; Vert, J.P.; Pollet, N.; Harland, R.M.; et al. Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Dev. Biol. 2014, 386, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Sasai, N.; Sasai, Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development 2005, 132, 2355–2363. [Google Scholar] [CrossRef] [Green Version]
- Simões-Costa, M.S.; McKeown, S.J.; Tan-Cabugao, J.; Sauka-Spengler, T.; Bronner, M.E. Dynamic and Differential Regulation of Stem Cell Factor FoxD3 in the Neural Crest Is Encrypted in the Genome. PLoS Genet. 2012, 8, e1003142. [Google Scholar] [CrossRef] [Green Version]
- Elms, P.; Scurry, A.; Davies, J.; Willoughby, C.; Hacker, T.; Bogani, D.; Arkell, R.M. Overlapping and distinct expression domains of Zic2 and Zic3 during mouse gastrulation. Gene Expr. Patterns 2004, 4, 505–511. [Google Scholar] [CrossRef] [Green Version]
- Diamand, K.E.M.; Barratt, K.S.; Arkell, R.M. Overview of Rodent Zic Genes; Springer: Singapore, 2018; Volume 1046. [Google Scholar]
- Furushima, K.; Murata, T.; Matsuo, I.; Aizawa, S. A new murine zinc finger gene, Opr. Mech. Dev. 2000, 98, 161–164. [Google Scholar] [CrossRef]
- Nagai, T.; Aruga, J.; Takada, S.; Günther, T.; Spörle, R.; Schughart, K.; Mikoshiba, K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol. 1997, 182, 299–313. [Google Scholar] [CrossRef]
- Houtmeyers, R.; Souopgui, J.; Tejpar, S.; Arkell, R.M. The ZIC gene family encodes multi-functional proteins essential for patterning and morphogenesis. Cell. Mol. Life Sci. 2013, 70, 3791–3811. [Google Scholar] [CrossRef]
- Inoue, T.; Hatayama, M.; Tohmonda, T.; Itohara, S.; Aruga, J.; Mikoshiba, K. Mouse Zic5 deficiency results in neural tube defects and hypoplasia of cephalic neural crest derivatives. Dev. Biol. 2004, 270, 146–162. [Google Scholar] [CrossRef]
- Elms, P.; Siggers, P.; Napper, D.; Greenfield, A.; Arkell, R.M. Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev. Biol. 2003, 264, 391–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, T.; Aruga, J.; Minowa, O.; Sugimoto, T.; Ohno, Y.; Noda, T.; Mikoshiba, K. Zic2 regulates the kinetics of neurulation. Proc. Natl. Acad. Sci. USA 2000, 97, 1618–1623. [Google Scholar] [CrossRef] [Green Version]
- Furushima, K.; Murata, T.; Kiyonari, H.; Aizawa, S. Characterization of Opr deficiency in mouse brain: Subtle defects in dorsomedial telencephalon and medioventral forebrain. Dev. Dyn. 2005, 232, 1056–1061. [Google Scholar] [CrossRef]
- Badis, G.; Berger, M.F.; Philippakis, A.A.; Talukder, S.; Gehrke, A.R.; Jaeger, S.A.; Chan, E.T.; Metzler, G.; Vedenko, A.; Chen, X.; et al. Diversity and complexity in DNA recognition by transcription factors. Science 2009, 324, 1720–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, L.S.; Hong, F.H.; Kunarso, G.; Stanton, L.W. The pluripotency regulator Zic3 is a direct activator of the Nanog promoter in ESCs. Stem Cells 2010, 28, 1961–1969. [Google Scholar] [CrossRef]
- Koyabu, Y.; Nakata, K.; Mizugishi, K.; Aruga, J.; Mikoshiba, K. Physical and functional interactions between Zic and Gli proteins. J. Biol. Chem. 2001, 276, 6889–6892. [Google Scholar] [CrossRef] [Green Version]
- Pourebrahim, R.; Houtmeyers, R.; Ghogomu, S.; Janssens, S.; Thelie, A.; Tran, H.T.; Langenberg, T.; Vleminckx, K.; Bellefroid, E.; Cassiman, J.J.; et al. Transcription factor Zic2 inhibits Wnt/Beta-catenin protein signaling. J. Biol. Chem. 2011, 286, 37732–37740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedard, J.E.J.; Purnell, J.D.; Ware, S.M. Nuclear import and export signals are essential for proper cellular trafficking and function of ZIC3. Hum. Mol. Genet. 2007, 16, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Hatayama, M.; Tomizawa, T.; Sakai-Kato, K.; Bouvagnet, P.; Kose, S.; Imamoto, N.; Yokoyama, S.; Utsunomiya-Tate, N.; Mikoshiba, K.; Kigawa, T.; et al. Functional and structural basis of the nuclear localization signal in the ZIC3 zinc finger domain. Hum. Mol. Genet. 2008, 17, 3459–3473. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, A.B.; Sinha, A.; Fan, V.B.; Cadigan, K.M. The Wnt Transcriptional Switch: TLE Removal or Inactivation? BioEssays 2018, 40, 1700162. [Google Scholar] [CrossRef] [PubMed]
- Fujimi, T.J.; Hatayama, M.; Aruga, J. Xenopus Zic3 controls notochord and organizer development through suppression of the Wnt/β-catenin signaling pathway. Dev. Biol. 2012, 361, 220–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Wang, L.; Bartom, E.; Marshall, S.; Rendleman, E.; Ryan, C.; Shilati, A.; Savas, J.; Chandel, N.; Shilatifard, A. β-Catenin/Tcf7l2–dependent transcriptional regulation of GLUT1 gene expression by Zic family proteins in colon cancer. Sci. Adv. 2019, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Aruga, J.; Kamiya, A.; Takahashi, H.; Fujimi, T.J.; Shimizu, Y.; Ohkawa, K.; Yazawa, S.; Umesono, Y.; Noguchi, H.; Shimizu, T.; et al. A wide-range phylogenetic analysis of Zic proteins: Implications for correlations between protein structure conservation and body plan complexity. Genomics 2006, 87, 783–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Ma, Y.; Qian, L.; Wang, J. Sumoylation regulates nuclear localization and function of zinc finger transcription factor ZIC3. Biochim. Biophys. Acta—Mol. Cell Res. 2013, 1833, 2725–2733. [Google Scholar] [CrossRef] [Green Version]
- Hendriks, I.A.; Vertegaal, A.C.O. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016, 17, 581–595. [Google Scholar] [CrossRef]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef] [Green Version]
- Jakobs, A.; Koehnke, J.; Himstedt, F.; Funk, M.; Korn, B.; Gaestel, M.; Niedenthal, R. Ubc9 fusion-directed SUMOylation (UFDS): A method to analyze function of protein SUMOylation. Nat. Methods 2007, 4, 245–250. [Google Scholar] [CrossRef]
- Jakobs, A.; Himstedt, F.; Funk, M.; Korn, B.; Gaestel, M.; Niedenthal, R. Ubc9 fusion-directed SUMOylation identifies constitutive and inducible SUMOylation. Nucleic Acids Res. 2007, 35, e109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamitani, T.; Nguyen, H.P.; Yeh, E.T.H. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J. Biol. Chem. 1997, 272, 14001–14004. [Google Scholar] [CrossRef] [Green Version]
- Bogani, D.; Warr, N.; Elms, P.; Davies, J.; Tymowska-Lalanne, Z.; Goldsworthy, M.; Cox, R.D.; Keays, D.A.; Flint, J.; Wilson, V.; et al. New semidominant mutations that affect mouse development. Genesis 2004, 40, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, J.N.; Ali, R.G.; Warr, N.N.; Wilson, H.M.; Bellchambers, H.M.; Barratt, K.S.; Thompson, J.; Arkell, R.M. A murine Zic3 transcript with a premature termination codon evades nonsense-mediated decay during axis formation. Dis. Model. Mech. 2013, 6, 755–767. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, J.N.; Diamand, K.E.M.; Bellchambers, H.M.; Arkell, R.M. Systematized reporter assays reveal ZIC protein regulatory abilities are Subclass-specific and dependent upon transcription factor binding site context. Sci. Rep. 2020, 10, 13130. [Google Scholar] [CrossRef]
- Poukka, H.; Aarnisalo, P.; Karvonen, U.; Palvimo, J.J.; Jänne, O.A. Ubc9 interacts with the androgen receptor and activates receptor- dependent transcription. J. Biol. Chem. 1999, 274, 19441–19446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizugishi, K.; Aruga, J.; Nakata, K.; Mikoshiba, K. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J. Biol. Chem. 2001, 276, 2180–2188. [Google Scholar] [CrossRef] [Green Version]
- Ishiguro, A.; Inoue, T.; Mikoshiba, K.; Aruga, J. Molecular properties of Zic4 and Zic5 proteins: Functional diversity within Zic family. Biochem. Biophys. Res. Commun. 2004, 324, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.Y.; Paraso, M.; Arkell, R.M.; Brown, S. In vitro analysis of partial loss-of-function ZIC2 mutations in holoprosencephaly: Alanine tract expansion modulates DNA binding and transactivation. Hum. Mol. Genet. 2005, 14, 411–420. [Google Scholar] [CrossRef]
- Munemitsu, S.; Albert, I.; Rubinfeld, B.; Polakis, P. Deletion of an amino-terminal sequence beta-catenin in vivo and promotes hyperphosporylation of the adenomatous polyposis coli tumor suppressor protein. Mol. Cell. Biol. 1996, 16, 4088–4094. [Google Scholar] [CrossRef] [Green Version]
- Ferrer-Vaquer, A.; Piliszek, A.; Tian, G.; Aho, R.J.; Dufort, D.; Hadjantonakis, A.K. A sensitive and bright single-cell resolution live imaging reporter of Wnt/ss-catenin signaling in the mouse. BMC Dev. Biol. 2010, 10, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewan, K.; Stubbs, M.; Todd, H.; Barbeau, O.; Botfield, H.; Young, R.; Ruddle, R.; Samuel, L.; Raynaud, F.; Allen, N.; et al. A Useful Approach To Identify Novel Small Molecule Inhibitors Of Wnt-Dependent Transcription. Cancer Res. 2011, 70, 5963–5973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, M.W.; Lee, S.; Furtado, M.B.; Xin, L.; Sparrow, D.B.; Martinez, C.G.; Dunwoodie, S.L.; Kurtenbach, E.; Mohun, T.; Rosenthal, N.; et al. Complex SUMO-1 Regulation of Cardiac Transcription Factor Nkx2-5. PLoS ONE 2011, 6, e24812. [Google Scholar] [CrossRef] [Green Version]
- Inoue, T.; Ota, M.; Mikoshiba, K.; Aruga, J. Zic2 and Zic3 synergistically control neurulation and segmentation of paraxial mesoderm in mouse embryo. Dev. Biol. 2007, 306, 669–684. [Google Scholar] [CrossRef] [Green Version]
- Kimura-Yoshida, C.; Mochida, K.; Ellwanger, K.; Niehrs, C.; Matsuo, I. Fate Specification of Neural Plate Border by Canonical Wnt Signaling and Grhl3 is Crucial for Neural Tube Closure. EBioMedicine 2015, 2, 513–527. [Google Scholar] [CrossRef] [Green Version]
- Nolan, P.M.; Peters, J.; Strivens, M.; Rogers, D.; Hagan, J.; Spurr, N.; Gray, I.C.; Vizor, L.; Brooker, D.; Whitehill, E.; et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat. Genet. 2000, 25, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Arkell, R.M.; Cadman, M.; Marsland, T.; Southwell, A.; Thaung, C.; Davies, J.R.; Clay, T.; Beechey, C.V.; Evans, E.P.; Strivens, M.A.; et al. Genetic, physical, and phenotypic characterization of the Del(13)Svea36H mouse. Mamm. Genome 2001, 12, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, N.; Ali, R.G.; Ahmed, J.N.; Arkell, R.M.; Voelkerding, K. High Resolution Melt Analysis (HRMA); a Viable Alternative to Agarose Gel Electrophoresis for Mouse Genotyping. PLoS ONE 2012, 7, e45252. [Google Scholar] [CrossRef]
- Barratt, K.S.; Arkell, R.M. Whole-Mount In Situ Hybridization in Post-Implantation Staged Mouse Embryos. Curr. Protoc. Mouse Biol. 2020, 10, e75. [Google Scholar] [CrossRef]
- Downs, K.M.; Davies, T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development 1993, 118, 1255–1266. [Google Scholar] [CrossRef]
- Barratt, K.S.; Arkell, R.M. Production of Digoxigenin-Labeled Riboprobes for In Situ Hybridization Experiments. Curr. Protoc. Mouse Biol. 2020, 10, e74. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.L.; Huang, C.J.; Chan, J.Y.H.; Liu, P.Y.; Chang, H.P.; Huang, S.M. Regulation of nuclear receptor and coactivator functions by the carboxyl terminus of ubiquitin-conjugating enzyme 9. Int. J. Biochem. Cell Biol. 2007, 39, 1035–1046. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellchambers, H.M.; Barratt, K.S.; Diamand, K.E.M.; Arkell, R.M. SUMOylation Potentiates ZIC Protein Activity to Influence Murine Neural Crest Cell Specification. Int. J. Mol. Sci. 2021, 22, 10437. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms221910437
Bellchambers HM, Barratt KS, Diamand KEM, Arkell RM. SUMOylation Potentiates ZIC Protein Activity to Influence Murine Neural Crest Cell Specification. International Journal of Molecular Sciences. 2021; 22(19):10437. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms221910437
Chicago/Turabian StyleBellchambers, Helen M., Kristen S. Barratt, Koula E. M. Diamand, and Ruth M. Arkell. 2021. "SUMOylation Potentiates ZIC Protein Activity to Influence Murine Neural Crest Cell Specification" International Journal of Molecular Sciences 22, no. 19: 10437. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms221910437