1. Introduction
Trypanosomatid parasites are protozoans that alternate between insect vectors and mammalian hosts to complete their life cycle [
1]. During their digenetic life cycle,
Leishmania are found as extracellular promastigotes in the intestinal tract of female sand flies [
2,
3] and as the obligatory intracellular amastigotes within the macrophages of mammalian hosts [
4]. This transition exposes the parasites to dramatic changes in their environmental conditions, which serve as signals to induce the developmental program of gene expression and stage differentiation. In higher eukaryotes, changes in environmental conditions induce a stress response that halts cap-dependent global protein synthesis while stress-response genes continue to be translated in a cap-independent manner [
5,
6]. However, exposure to the environmental switches described for
Leishmania are an integral part of the parasite life cycle.
As with other Trypanosomatids, regulation of protein coding genes in
Leishmania follows unique molecular features [
7,
8]. The protein-coding genes are arranged in large genomic clusters that are transcribed by RNA polymerase II to generate polycistronic mRNAs [
9,
10]. These mRNAs are further processed by trans-splicing and polyadenylation, to produce 5′capped and 3′poly (A) tailed mature mRNAs [
11,
12,
13]. In the absence of conventional mechanisms for transcriptional activation of protein-coding genes, mRNA processing, stability and translation drive the stage-specific program of gene expression [
8,
9,
14].
Translation initiation is a critical step in regulating protein synthesis in eukaryotes [
15]. The canonical pathway includes the formation of a pre-initiation complex that contains a cap-binding complex, eIF4F, that consists of an mRNA cap-binding protein eIF4E; a large scaffold protein eIF4G and an RNA helicase, eIF4A. The scaffold eIF4G recruits the poly (A) binding protein (PABP) to the 3′ poly (A) chain of the mRNA, leading to mRNA circularization [
16]. The association of the mRNA-bound eIF4F complex with other translation initiation factors together with the small ribosomal subunit complex scans the 5′ UTR until it reaches the first AUG, where translation initiates.
The
Leishmania genome encodes six paralogs of the cap-binding protein eIF4E and five paralogs of eIF4G [
17,
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29]. While the yeast genome encodes a single eIF4E that is essential for viability and two eIF4G homologs [
30,
31], mammalian cells encode for three eIF4Es and two eIF4Gs isoforms. The nematode
Caenorhabditis elegans encodes five eIF4E isoforms which possess variable affinities towards the m
7GTP and m
2,2,7GTP cap analogs [
32,
33]. Of the multitude of eIF4E isoforms in
C. elegans, not all play a role in canonical translation initiation and some are speculated to play a role as competitive inhibitors of eIF4G recruitment [
34].
The six eIF4Es of
Leishmania show poor sequence conservation with the eukaryotic eIF4Es or among themselves [
18]. This possibly reflects duplication events that occurred during evolution, adapting each paralog to cover discrete functions during the life cycle of the parasite [
35]. Analysis of the six
Leishmania eIF4E paralogs showed that LeishIF4E-4 binds to the cap analogs only during the promastigote stage and forms a canonical cap-binding complex with LeishIF4G-3 and LeishIF4A-1 [
18,
20,
21].
LeishIF4E-3 is a weak cap-binding protein but interacts with LeishIF4G-4 under normal conditions. However, during nutritional stress, also experienced during metacyclogenesis, the two proteins dissociate and LeishIF4E-3 enters into starvation-induced RNA granules [
36]. LeishIF4E-3 is essential in trypanosomatids and deletion of a single allele affected global translation and parasite infectivity, suggesting that LeishIF4E-3 has an active role in global protein synthesis under normal conditions [
25,
37].
LeishIF4E-2 is one of the least studied 4E paralogs in
Leishmania. It does not interact with any of the LeishIF4Gs, but migrates with the heavy polysome complexes in sucrose gradients, possibly stabilizing polysome complexes [
18]. Hemizygous deletion of LeishIF4E-2 does not affect global translation and growth rate but affects the morphology of promastigotes, possibly by changes in specific cytoskeleton proteins [
23]. Thus, LeishIF4E-2 does not appear to function as a general translation initiation factor and hence its function remains obscure.
The only LeishIF4E paralog that retains its cap-binding activity in axenic amastigotes is LeishIF4E-1. LeishIF4E-1 expression increases during metacyclogenesis and in amastigotes and maintains its cap-binding activity throughout these life stages. Although it does not interact with any of the LeishIF4G candidates in
Leishmania, it was shown to be involved in active translation [
38]. This was established by CRISPR-Cas9 mediated deletion that was marked by a reduction in global translation and changes in the promastigote specific proteome [
38]. The LeishIF4E-1 function is regulated by two LeishIF4E-1 interacting proteins, 4E-IP1 and 4E-IP2 [
21,
39]. The tight interaction of LeishIF4E-1 with its regulatory protein 4E-IP1 was verified by structural studies as well [
40].
Two additional isoforms of eIF4E have been identified in
Trypanosoma brucei, TbIF4E-5 and TbIF4E-6 [
41,
42]. Recently, we investigated the
Leishmania ortholog of TbIF4E-5. LeishIF4E-5 binds weakly to the m
7GTP cap analog, but only during the promastigote stage. Mass spectrometry analysis of proteins that co-purify with LeishIF4E-5 highlighted proteins involved in RNA metabolism, along with two LeishIF4G paralogs, LeishIF4G-1 and LeishIF4G-2. Overexpression of LeishIF4E-5 showed a decline in cell proliferation and an overall reduction in global translation. Thus, LeishIF4E-5 could be a putative translational repressor [
24].
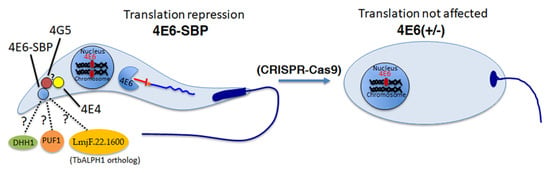
Here, we investigate LeishIF4E-6, the Leishmania ortholog of TbIF4E-6. We show that it has no affinity to the m7GTP cap analog, and in silico analysis provides a structural explanation to this observation. Overexpression of LeishIF4E-6 in transgenic parasites reduces global translation, excluding the possibility that LeishIF4E-6 is a canonical translation factor and possibly suggesting that it has a repressive role. The hemizygous deletion of LeishIF4E-6 affected the morphology of promastigotes, in a manner not yet understood.
3. Discussion
We focus on understanding the function of the different cap-binding protein paralogs of Leishmania and their role in translation. The recent development of CRISPR-Cas9 technology assists us with unraveling the function of these translation factors. In this study, we have characterized the function of the least studied cap-binding protein paralog, LeishIF4E-6 in Leishmania.
Sequence analysis highlights that the LeishIF4E-6 amino acid sequence is different from other LeishIF4Es and its mammalian counterpart and shows the highest similarity with LeishIF4E-5 (43%). Its degree of conservation with the T. brucei ortholog is also limited, reaching only 56% similarity. The unique sequences of the Leishmania cap-binding proteins which are different from their mammalian counterparts could be the consequence of evolutionary divergence.
LeishIF4E-6 overexpressing cells show a significant reduction in their global translation as compared with cells that overexpress LeishIF4E-1-SBP. Unlike LeishIF4E-6, LeishIF4E-1 has been demonstrated to be involved in translation activity and is required for
Leishmania growth and virulence [
38]. The global translation was evaluated using the SUnSET assay that monitors the incorporation of puromycin into nascent chains and stops their elongation. Although we saw that episomal expression of transgenes caused a reduction in total translation possibly due to competition for translation factors [
23,
38,
39], the reduction in translation in the LeishIF4E-6-SBP over-expression line was more significant as compared to controls. Therefore, global translation in the LeishIF4E-6 hemizygous knockout was less affected as compared to the LeishIF4E-6-SBP over-expressing line, although it was weaker than the Cas9/T7 control. The inhibitory effect of LeishIF4E-6 on translation activity and the lack of its m
7GTP binding are unique to
Leishmania and were not observed in the
T. brucei ortholog, TbIF4E-6. LeishIF4E-6(+/−) showed ~50% reduction in its mRNA level when compared to WT control. The strong decrease in LeishIF4E-6 RNA could have affected the protein level of LeishIF4E-6, resulting in the effects observed on global translation and cell morphology. In the LeishIF4E-6 hemizygous mutants, we noticed a phenotypic defect in the mutant promastigotes which became small and round with reduced flagellar length. However, the morphological changes observed in the LeishIF4E-6(+/−) mutant cells correlate with the changes in cell motility of
T. brucei cells, in which TbEIF4E-6 was silenced [
42]. Our results showing that the hemizygous mutant cells growing slower could be due to the potentially essential function of LeishIF4E-6, but the growth defects could also be caused by the altered morphology of the hemizygous mutant. In contrast, overexpression of LeishIF4E-6 did not affect cell morphology, even though its global translation was reduced. Although these results were unexpected, they could be explained if LeishIF4E-6 functions in a transcript-specific manner.
Although LeishIF4E-6 is classified as a cap-binding protein and its predicted structure indicates that it has a typical cap-binding pocket, we did not observe any m
7GTP-binding activities in any life cycle stage of
Leishmania. However, we cannot exclude the possibility that LeishIF4E-6 has limited cap-binding activity, below the resolution of our affinity chromatography assays. In any case, its inability to bind m
7GTP could suggest that LeishIF4E-6 may not be actively involved in translation initiation, supporting our observation of translation repression in LeishIF4E-6 overexpressing cells. The cap-binding pocket of the mammalian eIF4E consists of three tryptophan residues at positions 56, 102, and 166 of the mouse protein. These Trp residues are conserved in most eIF4Es but occasionally are replaced with other aromatic residues, such as Tyr or Phe. However, the substitution of conserved Trp residues may influence their binding of m
7GTP cap, as observed for LeishIF4E-5, where the Trp residue that is analogous to the mouse Trp102 is replaced with Tyr [
24]. Other residues may also assist the cap-binding activity, as basic residues within the pocket can interact with the acidic backbone of the cap structure. In the human eIF4E, Trp56 and Trp102 generate a π sandwich that stabilizes its binding to m
7GTP. In LeishIF4E-6, these amino acids are substituted with Tyr36 and Phe82, respectively. Although these substitutions maintain aromatic residues in these positions, these changes can lead to a reduction of the π interactions between Phe82 and Tyr36 with the m
7GTP, thus leading to a decreased affinity between LeishIFE-6 and m
7GTP. Unlike our observation that LeishIF4E-6 does not bind m
7GTP, the
T. brucei ortholog, TbIF4E-6, was reported to associate with m
7GTP. The different cap-binding activities observed for the
Leishmania and
Trypanosoma orthologs could originate from the altered residues that occupy the position which parallels Trp56 in the human eIF4E (Tyr in
Leishmania and Phe in
T. brucei).
Different eIF4E orthologs have been shown to form specific cap-binding complexes during the life cycle of
Leishmania and
Trypanosoma. We have identified the interacting proteins that are part of the LeishIF4E-6 complex using the pull-down analysis of tagged LeishIF4E-6. We noticed the enrichment of distinct classes of proteins including metabolic, transport-related, nuclear, mitochondrial, cytoskeletal, translation factors and chaperones that co-purified with LeishIF4E-6. Co-purification of chaperones was expected, as these are generally associated with overexpressed proteins. Among the translation factors, we observed a strong association of LeishIF4E-6 with LeishIF4G-5 and the G5-interacting protein (G5-IP). The LeishG5-IP contains the domains typically present in mRNA capping enzymes. Although the TbG5-IP was shown to be a homolog of the capping enzyme in
Crithidia fasciculata and
T. brucei, other proteins involved in the capping process were not present in the LeishIF4E-6 interactome [
49].
We also noted that the canonical translation factor LeishIF4E-4 co-purified with LeishIF4E-6 rather efficiently. The co-purification of LeishIF4E-4 with LeishIF4E-6 could indicate that components of the LeishIF4E-6 complex were able to sequester LeishIF4E-4, the canonical translation factor, preventing its binding to the 5′ cap structure of mRNAs. This could be one of the possible mechanisms explaining the translation repression activity of LeishIF4E-6, although this possibility requires further investigation. We also noted that LeishIF4G-3 that usually accompanies LeishIF4E-4 was not found in the LeishIF4E-6 interactome.
Enrichment of the proteins associated with ribonucleoprotein granules could suggest that LeishIF4E-6 interacts with RNA granule proteins that are involved in translation repression and mRNA degradation. We observed that the ATP-dependent RNA helicase DHH1 (LmjF.35.0370) was enriched in the LeishIF4E-6 associated proteins. DHH1 is known to regulate the balance between active translation, P-bodies formation and cytoplasmic decay of mRNAs [
50]. However, in trypanosomes, it is also known to play a selective role in determining the expression levels of developmentally regulated mRNAs [
51]. Furthermore, a
Leishmania homolog (LmjF.22.1600) of TbALPH1 was also found to be enriched in the LeishIF4E-6 interactome (
Table S1). In Trypanosomes, TbALPH1 is an mRNA decapping enzyme, identified in stress granules, and it also co-localizes with XRNA (5′-3′ exoribonuclease) in the posterior pole granules [
48]. Furthermore, we found that PUF1 (LmjF.36.0050), the Alba-domain protein 1 (LmjF.13.0450) and an RNA binding protein (LmjF.18.0590) co-purified with LeishIF4E-6. In
Leishmania and
T. brucei, PUF1 was reported to localize to cytoplasmic starvation-induced stress granules, which store mRNAs following their stalled translation [
36,
52]. In higher eukaryotes, PUF proteins are involved in translation repression either by blocking cap-binding events or by recruiting the deadenylating complex to remove the poly (A) tails [
53]. In trypanosomatids, the association of PUF1 with stress granules suggests that it could have a role in translation repression.
The
L. infantum homolog of the Alba-domain protein 1 (LiAlba1, LmjF.13.0450) is LINF_130009400 (previously Linj.13.0270). LiAlba1 and LiAlba3 were reported to form a complex with other RNA-binding proteins, ribosomal subunits, and translation factors and both proteins migrate to heavier sucrose fractions along with ribosomal proteins upon conditions inducing translational decline. This suggests a potential role of LiAlba1 in translational repression [
54]. The
L. infantum homolog of the RNA binding protein (LmjF.180590) LINF_180010900 (previously Linj.18.0590) was reported to interact with LiAlba1 only during the promastigote stage. LINF_180010900 is a 44 kDa RNA binding protein specific to
Leishmania [
54]. The association of PUF1 with LeishIF4E-6 shows that the latter could have a role in post-transcriptional RNA turnover [
55]. Enrichment of metabolic enzymes in the LeishIF4E-6 interactome could result from their moon-lighting RNA binding activity [
47].
The GO enrichment analysis highlighted the co-purification of proteins of the tethering complex group that have a primary function in the trafficking of proteins. These included orthologs of the conserved oligomeric complex 6, COG6 (LmjF.34.0370), that was proposed to function in membrane tethering during vesicular trafficking at the Golgi apparatus [
56]. Another protein, the exocyst complex component Exo99 (LmjF.21.1569) is an essential subunit of the nonameric exocyst complex first identified in
Trypanosoma brucei [
57]. In trypanosomes, Exo99 has a role in viability, morphology and trafficking as well as in maintaining the surface proteome. A TRAPPC-like protein (LmjF.16.1400) co-purifies with LeishIF4E-6. The subunit-like protein TRAPPC which in mammals is part of the trafficking protein particle complex (TRAPP complex), which mediates the contact between vesicles and target membranes [
58]. The GO enrichment analysis also identified components of the contractile vacuoles with LeishIF4E-6, based on the presence of calmodulin in the interactome. Contractile vacuoles are involved in the regulation of cell volume, by control of the water quantity within the cell and are involved in adaptation to high osmolarity conditions. The involvement of calmodulin in contractile vacuoles has been reported [
58], and calmodulin, which is enriched in the LeishIF4E-6 interactome, is rich in the membrane of contractile vacuole complexes. Calmodulin is a cytosolic calcium-binding messenger protein that mediates Ca2+ regulation in a wide range of cellular processes, such as activation of regulatory enzymes (e.g., adenylyl cyclases, ATPase) [
59] and reversible modulation of calcium-dependent processes [
60,
61]. Given the above, co-purification of contractile vacuole components with LeishIF4E-6 could possibly indicate why cell morphology is altered in the LeishIF4E-6(+/−) mutant [
62,
63], although other mechanisms for this effect cannot be ruled out.
We identified LeishIF4G-5 as a binding partner of LeishIF4E-6 through mass spectrometry analysis of proteins that were pulled down with LeishIF4E-6. The association between LeishIF4G-5 and LeishIF4E-6 was verified in vitro, by monitoring the interaction between LeishIF4E-6 and recombinant LeishIF4G-5. The interaction between the two proteins was also observed in
T. brucei, suggesting that they form an evolutionarily conserved complex. The YXXXXLϕ domain that is found in eIF4E-binding proteins is also shared by the 4E-interacting proteins in
Leishmania [
21,
39]. Sequence analysis of LeishIF4G-5 identified two conserved putative YXXXXLϕ domains, one at the center of the protein and another at the C-terminus of LeishIF4G-5. However, the latter element has a cysteine residue at its 6th position of the suggested 4E-binding domain. It should be noted though that analysis of the interaction between LeishIF4G-3 and LeishIF4E-4 showed that the YXXXXLϕ domain in LeishIF4G-3 was only partially conserved, as its 6th position is occupied by Aspartic acid, rather than by a Lysine residue [
20]. Thus, the suggested 4E-binding domain at that C-terminus of LeishIF4G-5 could be functional. The presence of two 4E-binding motifs in LeishIF4G-5 could also suggest that this protein is capable of binding to both LeishIF4E-6 and LeishIF4E-4. The co-purification of LeishIF4E-4 with LeishIF4E-6 could be mediated by other specific proteins in the LeishIF4E-6 complex, which were not yet identified. The presence of LeishIF4E-4 in this complex could therefore potentially bring LeishIF4E-4 bound transcripts to interact with the LeishIF4E-6 bound proteins that are involved in RNA inactivation such as TbALPH1 homolog, PUF1 and DHH1. Further investigations are required to establish the interaction between LeishIF4G-5 and LeishIF4E-4. We also observed that LeishIF4G-5 is subjected to proteolysis. To confirm that the cleavage products were derived from LeishIF4G-5, we carried out the mass spectrometry of each band detected in Western blots and found LeishIF4G-5 specific peptides. The biological role of this cleavage is still not clear.
Overall, we report that LeishIF4E-6 is a unique protein that does not associate with the m7GTP cap. Its inability to interact with the mRNA cap suggests that LeishIF4E-6 could not promote translation initiation, possibly suggesting that LeishIF4E-6 could have a role in translation repression, as suggested by the global translation assays. The interactome of LeishIF4E-6 is rich with proteins involved in RNA inactivation such as DHH1, the TbALPH1 homolog, PUF1 and the Alba-domain protein 1. These proteins have an established role in translation repression through mRNA degradation (ALPH1 homolog) or via sequestering of mRNAs to dedicated storage granules (DHH1, PUF1). Although LeishIF4E-6 does not directly interact with the LeishIF4E-4, its interaction with LeishIF4G-5 could tether LeishIF4E-4 to the LeishIF4E-6 complex. Such an interaction could potentially sequester LeishIF4E-4 to dedicated foci that contain enzymes that have an overall inhibitory effect on translation, such as DHH1, PUF1, and the Alba-domain protein 1. Moreover, the presence of the decapping enzyme homolog of TbALPH1 in the complex could further support the inhibitory nature of LeishIF4E-6 on translation. This could be a potential mechanism by which LeishIF4E-6 regulates protein translation. The requirement for the LeishIF4E-6 and LeishIF4G-5 complex formation presents a unique and intriguing regulatory mechanism that will be further investigated.
4. Materials and Methods
4.1. Organisms and Cell Culture
Leishmania amazonensis (MHOM LTB0016) and Leishmania mexicana (M379) promastigotes were cultured at 25 °C in Medium 199 [M199, (pH 7.4)] (Biological Industries, Beit Haemek, Israel) containing 10% fetal calf serum (FCS, Biological Industries, Beit Haemek, Israel), 5 µg/mL hemin, 0.1 mM adenine (Sigma, St. Louis, MO, USA), 40 mM HEPES (Sigma, St. Louis, MO, USA), 4 mM L-glutamine (Biological Industries, Beit Haemek, Israel), 100 U/mL penicillin and 100 µg/mL and streptomycin (Biological Industries, Beit Haemek, Israel). Leishmania amazonensis axenic amastigotes were generated using promastigote cells from late log phase (3.6 × 107 cells/mL) which were washed twice with phosphate-buffered saline (PBS) and resuspended in M199, containing 25% FCS, 5 µg/mL hemin, 0.1 mM adenine, 40 mM HEPES pH 5.5 (adjusted by using 0.5 M succinic acid), 4 mM L-glutamine, 100 U/mL penicillin and 100 µg/mL streptomycin. After resuspension in axenic amastigote specific media, the cells were further grown at 33° C for four days with constant shaking.
4.2. Generation of Transgenic Leishmania Overexpressing the SBP Tagged LeishIF4E-6
The LeishIF4E-6 open reading frame (ORF, 549 bp) was amplified from
L. amazonensis genomic DNA using gene-specific primers, and cloned into a pX-derived transfection cassette, pX-H-target ORF-H-SBP between the BamHI/XbaI sites. In this transfection, cassette H represents the intergenic region of HSP83 derived from the genomic locus [
21] of
Leishmania and SBP (streptavidin-binding peptide, 6 kDa) is the C-terminal tag. Mid-log phase
L. amazonensis cells (50 mL) were transfected with 40 μg of the construct, pX-H-LeishIF4E-6-SBP-H- following a published protocol [
64]. The positive clones were selected for resistance to G418 (200 μg/mL).
4.3. Sequence Analysis of LeishIF4E-6
A multiple sequence alignment was carried out and the secondary structure of LeishIF4E-6 was predicted. The ORFs of LeishIF4E-6 from various
Leishmania and Trypanosoma species along with mammalian and
L. amazonensis LeishIF4E-1 were aligned using the Jalview (2.11.1.4) alignment tool [
65]. The sequences used were
Mus musculus eIF4E-1 (ENSMUSG00000028156),
L. amazonensis LeishIF4E-1 (LAMA_000544700),
L. amazonensis LeishIF4E-6 (LAMA_000504800);
L. mexicana LeishIF4E-6 (LmxM.26.0240)
L. donovani LeishIF4E-6 (LdBPK.26.2.000230), and
T. brucei TbIF4E-6 (Tb927.7.1670). The alignment file that was generated with Jalview was first saved in FASTA format. Then, the PDB file (6YLT, doi:10.2210/pdb6YLT/pdb) of
Mus musculus eIF4E-1 was downloaded from the protein database (
https://www.rcsb.org/structure/6YLT, accessed on 5 July 2021). The final sequence and structural alignment showing the predicted secondary structure was developed using the downloaded PDB file (6YLT, doi:10.2210/pdb6YLT/pdb) of
Mus musculus eIF4E-1 along with the FASTA alignment file, using the online ESPript 3 tool ESPript 3 (ibcp.fr). The separate alignment
L. amazonensis of LeishIF4E-6 and
T. brucei TbIF4E-6 were also carried out using Jalview 2.11.1.4, to highlight the homology between these two sequences. Sequence similarities of LeishIF4E-6 with various LeishIF4Es from
Leishmania,
Mus musculus eIF4E1 and
Trypanosoma TbIF4E-6 were generated by the EMBOSS needle tool (
https://www.ebi.ac.uk/Tools/psa/emboss_needle/, accessed on 6 May 2021). Similarly, sequence similarities of LeishIF4G-5 with various LeishIF4Gs from
Leishmania and
Mus musculus eIF4G were also generated.
4.4. Structural Homology Modeling of LeishIF4E-6
Structural homology modeling was performed on LeishIF4E-6 via SWISS-MODEL workspace [
65]. Several modeling structures were chosen and superposed on their known template structures using WinCoot 0.9.4.1 [
66] and the best template structure was chosen based on the quality of the solved structures, the presence of ligand, and the completeness and the orientation of the pocket loops. Images were prepared via ChimeraX 1.2.4 and GIMP 2.10.24 used for the graphical editing.
4.5. Confocal Microscopy to Monitor the Localization of LeishIF4E-6-SBP
L. amazonensis promastigotes overexpressing LeishIF4E-6-SBP were examined by confocal microscopy. The transgenic cells were cultured in 8 well μ-Slide plates (Ibidi GmbH, Gräfelfing, Germany) which fits in the microscope chamber, and mid-log cells (~107 cells/mL) were processed for imaging. The cells were washed once in serum-free M199 media and then fixed in 2% paraformaldehyde in PBS for 20 min at room temperature and then washed with PBS once. Cells were permeabilized with 0.1% Triton X-100 in PBS, washed again with PBS and blocked with 2% bovine serum albumin (BSA) in PBS at room temperature for 1 h. The fixed cells were then incubated with a monoclonal antibody against the SBP tag (1:100) (Millipore, Ternecula, CA, USA) for an hour at room temperature. The cells were washed again with PBS and incubated with and a secondary anti-mouse IgG DyLight 488 (3:500) (KPL, Milford, CT, USA) for an hour at room temperature. The nuclear and kinetoplast DNA was stained using 4′, 6-diamidino-2-phenylindole (DAPI; 1 μg/mL of PBS; Sigma). An inverted Zeiss LSM 880 Axio-observer Z1 confocal laser-scanning microscope with Airyscan detector was used to capture the images. A Plan-Apochromat 63x/1.4 oil, 1.4 Numeric aperture (NA), differential Interference Contrast (DIC) objective was used to visualize the cells. Images were recorded at a 512 x 512 pixels format along with 8x digital zoom for an enlarged view of
Leishmania promastigote. Airyscan processing of the images was carried out using Zen lite software (Carl Zeiss AG, Oberkochen, Germany). ImageJ software was further used to generate a single representative section of Z-projection with maximum intensity [
67].
4.6. Monitoring Global Translation Activity
Global translation in transgenic
L. amazonensis promastigotes overexpressing LeishIF4E-6-SBP along with control wild type (WT) cells and cells overexpressing the transgenic LeishIF4E-1-SBP and the chloramphenicol acetyltransferase (CAT) reporter protein (flanked by the HSP83 intergenic regions (i), pX-iCATi, was monitored using the SUrface SEnsing of Translation (SUnSET) assay. This assay is based on the incorporation of puromycin, a structural tRNA analog, into the growing polypeptide in the A site of the translating ribosome [
68]. Quantification of puromycin incorporated serves as an indication for global translation in the examined cells. The different cell lines described above were incubated with 1 µg/mL of puromycin (Sigma, St. Louis, MO, USA) for 30 min, washed twice with PBS and once with PRS
+ buffer (PRS: 35 mM HEPES, 100 mM KCl, 10 mM MgCl
2,1 mM dithiothreitol, DTT; PRS
+: PRS supplemented with Phosphatase inhibitors [20 mM NaF, 50 mM β-glycerophosphatase] and Protease inhibitors [2× protease inhibitors cocktail, 2 mM iodoacetamide]). The washed cell pellets were resuspended in 300 µL of PRS
+ buffer and Laemmli sample buffer (2×) and boiled for 5 min. Cells treated with Cycloheximide (100 μg/mL) (Sigma, St. Louis, MO, USA) prior to the addition of puromycin served as the negative control for translation. The samples were resolved over 10% SDS- SDS-polyacrylamide (PAGE) gels. Western analysis was carried out using specific primary monoclonal antibodies against puromycin (DSHB, University of Iowa, Iowa, USA; 1:1000) and secondary peroxidase-labeled anti-mouse antibodies (KPL, Milford, CT, USA; 1:10,000)
4.7. Affinity Purification over m7GTP-Agarose
Transgenic lines overexpressing the SBP-tagged LeishIF4E-6 and LeishIF4E-1 (as a positive control) were used in the assay. The cells (∼109) were washed twice with PBS and once with column buffer, (CB, 20 mM HEPES, pH 7.4, 2 mM EDTA, 1 mM DTT and 50 mM NaCl). The cell pellets were resuspended in 1.2 mL of CB+ which consisted of CB containing protease and phosphatase inhibitors (a commercial mix of protease inhibitors (Sigma), 4 mM iodoacetamide, 25 mM sodium fluoride and 55 mM β-glycerophosphate). Leishmania cells were lysed with 1% Triton X-100 in CB+ on ice for 5 min. The supernatants were clarified by centrifugation at 20,000× g for 20 min at 4 °C. The clarified supernatants were then incubated for 2 h with m7GTP -agarose resin (75 μL) (Jena Biosciences, Jena, Germany) that were pre-equilibrated with CB. The m7GTP -agarose resins bound to protein complexes were washed with CB twice and once with CB supplemented with 100 μM GTP. Finally, the elution of the cap-binding complexes was carried out with CB+ containing 200 μM of free m7GTP. The eluted fractions were precipitated with trichloroacetic acid (TCA), at a final concentration of 10% at 4 °C overnight under constant shaking. The proteins were spun down at 20,000× g for 20 min at 4 °C and the pellets were washed with 100% chilled acetone, briefly dried and resuspended in Laemmli sample buffer. Aliquots derived from the supernatant (5%), the flow-through (5%), wash (50%) and elution (50%) fractions were resolved over 10% SDS-PAGE gels, blotted and western analysis was carried out with specific monoclonal antibodies against SBP tag.
4.8. Affinity Purification over Streptavidin-Sepharose Beads
Lysates of cells overexpressing SBP-tagged LeishIF4E-6 and Luciferase were affinity-purified over streptavidin-Sepharose beads. The cells (~109) were washed twice with PBS, once with PRS, then lysed with 1% Triton X-100 in 1.2 mL PRS+ for 5 min in ice. The cell lysates were clarified by centrifugation at 20,000× g for 20 min at 4 °C. The clarified supernatants (1.2 mL each) were then incubated for 2 h with 75 μL streptavidin-Sepharose beads (GE Healthcare, Buckinghamshire, UK) pre-equilibrated with PRS buffer for 2 h. After collecting the flow-through, the beads were washed three times with PRS+ containing 0.1% NP-40. The final elution was carried out with 5 mM biotin in PRS+ supplemented with 0.1% NP-40. The eluted fractions were precipitated with trichloroacetic acid (TCA) at a final concentration of 10% at 4 °C overnight with constant shaking. The proteins were precipitated by centrifugation at 20,000× g for 20 min at 4 °C washed with 100% chilled acetone and finally resuspended in Laemmli sample buffer. Aliquots of the different fractions including the supernatant (5%), the flow-through (5%), the final wash (50%) and the elution (50%) were separated over 12% SDS-PAGE gels, blotted and western analysis was carried out with specific antibodies. The eluted samples from the SBP-tagged LeishIF4E-6 and Luciferase cell lines were then subjected to mass spectrometry analysis.
4.9. Proteomic Analysis by Mass Spectrometry
Lysates of cells overexpressing SBP-tagged LeishIF4E-6 and Luciferase were affinity-purified over streptavidin-Sepharose beads. The eluted proteins were precipitated with 10% TCA, and the pellets were washed with 100% chilled acetone. The protein pellets were resuspended in Laemmali’s buffer and further separated over 12% SDS-PAGE gels that were stained with Coomassie Brilliant Blue. The relevant lanes were excised and subjected to mass spectrometry analysis in the Smoler Proteomics Center at the Technion, Israel.
Mass Spectrometry-Proteins were reduced by adding 3 mM DTT at 60 °C for 30 min. This was followed by modification with 10 mM iodoacetamide in 100 mM ammonium bicarbonate (Promega, Madison, WI, USA) (30 min at room temperature). The proteins were then digested with trypsin (Promega) in 10 mM ammonium bicarbonate at 37 °C overnight. The trypsin-digested peptides were desalted, dried, and resuspended in 0.1% formic acid and finally resolved by the reverse phase chromatography over a 30 min linear gradient of 5% to 35% acetonitrile and 0.1% formic acid in water, a 15 min gradient of 35% to 95% acetonitrile and 0.1% formic acid in water and a 15 min gradient of 95% acetonitrile and 0.1% formic acid in water at a flow rate of 0.15 µL/min. Mass spectrometry (MS) was carried out using a Q Exactive Plus mass spectrometer (Thermo Fischer Scientific, Waltham, MA, USA) in the positive mode set to conduct a repetitively full MS scan followed by the high energy collision dissociation of the 10 dominant ions selected from the first MS scan. A mass tolerance of 10 ppm for precursor masses and 20 ppm for fragment ions was set.
Statistical Analysis—MaxQuant software (version 1.5.2.8) [
69] was used to analyze the raw data generated by mass spectrometry to identify the proteins that were identified in the lysates of cells overexpressing LeishIF4E-6 as compared to the luciferase-SBP control The raw data were searched against the highly annotated L. major Friedlin strain proteins enlisted in the TriTrypDB database [
46]. Protein identification parameters were set at less than a 1% false discovery rate. The MaxQuant parameter settings included a minimum of 1 razor/unique peptide for identification, a minimum peptide length of six amino acids and a maximum of two mis-cleavages. Summed peptide intensities were used for the protein quantification. Missing intensities of the proteins in control Luciferase samples were substituted with values close to the baseline. To identify the proteins that were enriched in the LeishIF4E-6 interactome, the log
2 of iBAQ intensities [
70] were compared between three LeishIF4E-6-SBP biological repeats and three luciferase- SBP repeats on the Perseus software platform [
71], using the
t-test. Adjusted
p-values were corrected using permutation-based false discovery rate (FDR) = 0.05 and the number of randomizations = 250. These are marked as Padj [
71]. The enrichment threshold was set to a log
2 fold change > 1.6 and a
p-value < 0.05. The annotated proteins were first categorized manually based on the functions. Proteins enriched in the LeishIF4E-6 as compared to Luciferase-SBP were manually categorized into different groups based on their functions and represented by the pie chart.
4.10. Gene Ontology (GO) Annotation of Enriched Proteins
The proteins that were enriched in the LeishIF4E-6-SBP interactome as compared to the control Luciferase-SBP were further annotated by the GO Annotation tool in TriTrypDB, based on the cellular components’ parameter. The threshold for this analysis of proteins based on their GO terms was set at four-fold, with a p-value < 0.01. This threshold eliminated most of the general groups that represented parental GO terms. GO terms based on only a single protein were also filtered out.
4.11. Cloning and Expression of Recombinant GST-Tagged LeishIF4E-6
LeishIF4E-6 (549 bp) ORF was amplified by PCR using
L. amazonensis genomic DNA with the specific set of forward primer 5′-gctgaattcATGGCCGATAGTAATCCCACCA-3′ and reverse primer 5′-gctctagaCTA CTT GAA GGG GCG AGC GCT-3′. Lower case letters represent restriction sites EcoRI in forward primer and XbaI in reverse primer while upper case letters are the sequences from LeishIF4E-6 ORF. The pGST-parallel expression plasmid was used for cloning of LeishIF4E-6 with an N-terminus GST tag [
72]. BL21
E. coli cells were transformed with the plasmid clone containing GST-tagged LeishIF4E-6. The BL21
E. coli cells were grown at 37 °C until OD
600 = 0.5, when expression was induced by the addition of 0.5 mM Isopropyl-β-d-1-thiogalactopyranoside (IPTG).
E. coli cells were further incubated for 14–16 h at 18 °C after IPTG addition.
4.12. Interaction between Recombinant GST-LeishIF4E-6 and LeishIF4G-5-SBP
Transgenic L. amazonensis cells overexpressing the SBP-tagged LeishIF4G-5 and BL21 E. coli cells expressing the recombinant GST-tagged LeishIF4E-6 were used for the interaction assay. Mid log phase Leishmania cells (~8× 106 cells/m) overexpressing the SBP tagged LeishIF4G-5 were washed with PBS twice and once with disruption buffer (DB, 20 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 5 mM MgCl2 and 5% glycerol). The cell pellets were then resuspended in the same buffer (DB) containing a cocktail of protease inhibitors (Sigma) and 2 mM iodoacetamide, along with phosphatase inhibitors (20 mM sodium fluoride, 50 mM β-glycerophosphatase) and 0.1% NP-40 (DB+). Cell lysis was carried out with 1% Triton X-100 for 5 min on ice. Clarified supernatant was further obtained by subjecting the lysates to centrifugation at 20,000× g for 20 min at 4 °C. Clarified supernatant containing SBP-tagged LeishIF4G-5 was then incubated with pre-equilibrated (with DB+ buffer) streptavidin-Sepharose beads (75 μL, GE Healthcare) for 2 h at 4 °C with constant shaking. The beads were then washed three times with DB+. Simultaneously, IPTG induced BL21 E. coli cells (200 mL) expressing the GST-tagged LeishIF4E-6 were washed twice with PBS, and once with DB. The BL21 E. coli cell pellets were resuspended in DB+ and disrupted in a French Press apparatus at 1500 twice. The disrupted cells were centrifuged at 45,000× g rpm in a Ti70 rotor (Beckman Coulter) for 45 min. The clarified bacterial supernatant containing the GST-tagged LeishIF4E-6 (10 mL) was then incubated with the streptavidin-Sepharose beads that were pre-bound with the SBP tagged LeishIF4G-5. This incubation was set for 2 h at 4 °C with constant shaking. After this incubation, the flow-through fraction was collected and the beads were washed five times with 1 mL of DB+. The final elution was carried out using 5 mM of biotin in 1 mL DB+. Aliquots derived from the supernatant (S, 2%), flow-through (FT, 2%), wash (W, 25%) and elution (E, 25%) fractions were separated over 12% SDS-PAGE and subjected to western analysis using antibodies against the SBP tag to detect LeishIF4G-5 (Millipore, Temecula, CA, USA). Antibodies against the GST tag (Invitrogen 1:1000) were used to detect LeishIF4E-6.
4.13. CRISPR-Cas9-Mediated Generation of a Hemizygous LeishIF4E-6 Mutant
The different plasmids optimized for the CRISPR-Cas9 mediated gene editing in
Leishmania were a kind gift from Eva Gluenz (University of Oxford, Oxford, UK) [
73]. The pTB007 plasmid was used to generate a
Leishmania mexicana cell line that expressed Streptococcus pyogenes CRISPR-associated protein 9 endonuclease and the T7 RNA polymerase gene (Cas9/T7). This cell line was selected for hygromycin resistance (200 μg/mL).
A LeishIF4E-6 hemizygous mutant was generated using different PCR-amplified products. The two 5′ and 3′ sgRNAs were derived from the respective untranslated region of LeishIF4E-6, to target the double-strand breaks. Other PCR products included the two different repair cassette fragments that introduced either the G418 or the blasticidin marker into the double-strand breaks in the genome. All four PCR products were electroporated into mid-log-phase transgenic cells overexpressing Cas9 and the T7 RNA polymerase. Cells were further selected for resistance to 200 μg/mL G418 [
38,
73], or 20 μg/mL Blasticidin or both. The sgRNA sequences that were used to delete the LeishIF4E-6 gene were obtained from
http://leishgedit.net (5 July 2020).
PCR Amplification of sgRNA Templates. LeishIF4E-6 specific 5′ and 3′ guide RNAs fragments for cleavage upstream and downstream of the LeishIF4E-6 target gene were generated by PCR. The template for this PCR included two partially complementary long oligonucleotides; the common sgRNA scaffold oligonucleotide (5′-AAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′), and another oligonucleotide that included the T7 RNA polymerase promoter (lowercase letters at the beginning of the sequence) fused to the gRNA (5′ or 3′) targeting LeishIF4E-6 (capital letters) and a short sequence overlapping the scaffold fragment (lowercase letters at the end of the sequence). The two individual template oligonucleotides designed for targeting the double-strand breaks at the 5′ and 3′ ends of LeishIF4E-6 were 5′-gaaattaatacgactcactataggCCGACTCGTCTGCGTAGCCGgttttagagctagaaatagc-3′ and 5′-gaaattaatacgactcactataggTGCGCATATGTTCATCAGTGgttttagagctagaaatagc-3′, respectively. These two template fragments (1 μg each) were annealed to their partially complementary sequence and further amplified with a pair of small primers (2 μM each) derived from the T7 promoter (G00F, 5′-TTAATACGACTCACTATAGG-3′) and the common scaffold fragment (G00R, 5′-GCACCGACTCGGTGCCACTT-3′). PCR conditions were described in [
38]. All PCR products were gel extracted and sterilized at 94 °C for 5 min before transfection.
PCR Amplification of the LeishIF4E-6 Replacement Fragment. The LeishIF4E-6 replacement fragment was amplified by PCR using the pTNeo plasmid as a template. The primers were (5′-CGCCACCGTTCCGCCGGCTTCCTTCAGCCTgtataatgcagacctgctgc-3′ [forward]) and 5′-ACACTCTATACACACATGCGCACACACGTAccaatttgagagacctgtgc-3′ [reverse]). The capital letters represent 5′ and 3′ UTR sequences in the Forward and Reverse primers, respectively. The lowercase letters represent the region on the pTNeo plasmid that flanks the UTR adjacent to the antibiotic resistance gene (G418 or Blasticidin). These primer pairs were designed based on the LeishGEdit database (
http://www.leishgedit.net/Home.html). The LeishIF4E-6 replacement fragment consisted of G418 sequences flanked by LeishIF4E-6 specific UTRs that promote the integration of the drug resistance marker by homologous recombination at the target site. PCR conditions were described in [
38]. All PCR products were gel extracted and heated at 94 °C for 5 min before electroporation in
Leishmania cells.
Diagnostic PCR to Confirm the Deletion of LeishIF4E-6. Genomic DNA was isolated from the
Leishmania cells transfected with different PCR products to knock out the LeishIF4E-6 gene after 14 days of G418 drug selection by using DNeasy Blood & Tissue Kit (Qiagen, Germantown, MA, USA). We carried out several diagnostic PCRs to detect the deletion of the LeishIF4E-6 gene, using different primer pairs. The first PCR included primers derived from LeishIF4E-6 UTRs, a forward primer from the 5′UTR (P1: 5′-CACAGCATCTCCATTCCTTCTT-3′) and a reverse primer derived from the 3′ UTR of LeishIF4E-6 (P2: 5′-TCTCCACCACCACGGAAT-3′). Another PCR reaction was performed to analyze the insertion of the G418 resistance gene using the G418 ORF primers which were G418 Forward (P3: 5′-GCCCGGTTCTTTTTGTCAAGAC-3′) and G418 Reverse (P4: 5′-GTCACGACGAGATCATCATCGCCG-3′). Another PCR reaction was carried out to confirm the insertion of the G418 cassette at the target site. This PCR was performed with primers derived from the G418 ORF (Forward P3) and the LeishIF4E-6 3′UTR (Reverse P2). Genomic DNA from the positive control Cas9/T7 overexpressing
L.
mexicana cells was used to detect the presence of the LeishIF4E-6 gene using the primer set derived from the 5′ and 3′ UTRs. The PCR conditions were described in [
38].
4.14. Quantitative Real-Time PCR
Total RNA was isolated from L. amazonensis promastigotes overexpressing LeishIF4E-6-SBP, along with control wild-type (WT) cells and cells overexpressing the transgenic LeishIF4E-1-SBP and the chloramphenicol acetyltransferase (CAT) reporter protein. Total RNA was also extracted from L. mexicana promastigotes overexpressing Cas9/T7, along with control wild type (WT) cells and L. mexicana 4E6+/− hemizygous mutant. The RNA was extracted from day 2 cells using the TRI reagent (Sigma), following the manufacturer’s instructions. The purified RNA was subjected to DNAse treatment by incubation at room temperature for 20 min followed by the heat inactivation of the DNAse (Thermo Fisher Scientific, Bleiswijk, Netherlands) at 65 °C for 10 min. Finally, the RNA samples were stored at −80 °C.
cDNA was synthesized from 1 μg total RNA from different cell lines in a 20 μL reaction mixture by the use of High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The cDNA was diluted 1:5 and 1 µL was used for the RT-PCR reaction. Reverse transcription reaction conditions included 25 °C for 10 min, 37 °C for 120 min, and 85 °C for 5 min followed by cooling at 4 °C. The real-time PCR reactions were carried out using a Applied Biosystem (cat # 4376600, StepOnePlus Real-Time PCR System) platform using the Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). Reaction mixtures included 5 μL 2X Fast SYBR Green Master Mix, 1 μL of each forward and reverse primers (5 pm each), 2 μL nuclease-free H2O and 1 μL cDNA. The primer sequences used for LeishIF4E-6 genes were FP (GAACGACGACAACCTCGTCT) and RP (TTGCAGGCGTCACGACTAAT). The primer sequences used for the endogenous control gene GAPDH were FP (GGGTAAGCTCGGTGTGGATTAC) and RP (CTGGTTCACACCCATCACGA). The PCR conditions were: 95 °C for 20 sec for initial activation followed by 40 cycles of 95 °C for 3 sec, and 60 °C for 30 sec. The PCR reaction was followed by a melting curve analysis to check the purity of the product. This was done at 95 °C for 15 sec, 60 °C for 60 sec and 95 °C for 15 sec. QRT-PCR reactions were analyzed using StepOne™ Software provided with the instrument. The relative expression of the genes to check fold differences was calculated by using the 2−ΔΔCT formula where endogenous control GAPDH was used as the normalizer. The values of three replicates were represented as a dot plot graph. The statistical analysis was carried out using GraphPad Prism 5 and significant differences were estimated by non-parametric T-test using a paired t-test.