Template-Assisted Plasmonic Nanogap Shells for Highly Enhanced Detection of Cancer Biomarkers
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Interior Nanogap-Containing Ag NGSs
2.2. Structural Analysis of the Time-and RLC-Dependent Formation of Ag NGSs
2.3. Characterization of SERS Sensitivities at the Single Particle Level and in Solution
2.4. Immunoassay Using Magnetic Beads (MBs) for PSA Detection
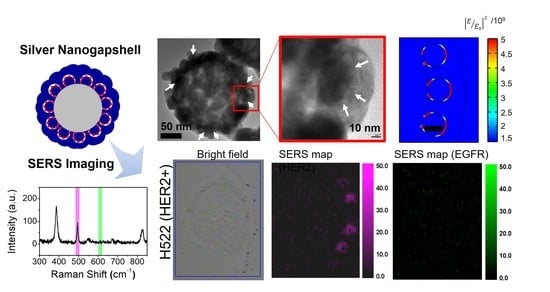
2.5. Cellular SERS Imaging of NSCLC Cells
3. Materials and Methods
3.1. Materials
3.2. Synthesis
3.2.1. Preparation of Ag NGSs
3.2.2. Silica Encapsulation of Ag NGSs and Antibody Conjugation for Immunoassays
3.2.3. Surface Modification of Ag NGSs for Extracellular Biomarker Detection
3.3. Theoretical Calculation of the E-field Enhancement of Ag NGS
3.4. SERS Measurements and Single-Particle Identification
3.5. Sandwich PSA Immunoassay Using Ag NGS Nanoprobes
3.6. Cell-Based Assay Using Ag NGS Nanoprobes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zong, C.; Xu, M.; Xu, L.J.; Wei, T.; Ma, X.; Zheng, X.S.; Hu, R.; Ren, B. Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef]
- Pallaoro, A.; Braun, G.B.; Moskovits, M. Biotags Based on Surface-Enhanced Raman Can Be as Bright as Fluorescence Tags. Nano Lett. 2015, 15, 6745–6750. [Google Scholar] [CrossRef]
- Cheng, Z.; Choi, N.; Wang, R.; Lee, S.; Moon, K.C.; Yoon, S.Y.; Chen, L.; Choo, J. Simultaneous Detection of Dual Prostate Specific Antigens Using Surface-Enhanced Raman Scattering-Based Immunoassay for Accurate Diagnosis of Prostate Cancer. ACS Nano 2017, 11, 4926–4933. [Google Scholar] [CrossRef]
- Chang, H.; Kang, H.; Yang, J.K.; Jo, A.; Lee, H.Y.; Lee, Y.S.; Jeong, D.H. Ag shell-Au satellite hetero-nanostructure for ultra-sensitive, reproducible, and homogeneous NIR SERS activity. ACS Appl. Mater. Interfaces 2014, 6, 11859–11863. [Google Scholar] [CrossRef]
- Kang, H.; Koh, Y.; Jeong, S.; Jeong, C.; Cha, M.G.; Oh, M.; Yang, J.-K.; Lee, H.; Jeong, D.H.; Jun, B.-H.; et al. Graphical and SERS Dual-Modal Identifier for Encoding OBOC Library. Sens. Actuators B Chem. 2020, 303, 127211. [Google Scholar] [CrossRef]
- Koo, K.M.; Wang, J.; Richards, R.S.; Farrell, A.; Yaxley, J.W.; Samaratunga, H.; Teloken, P.E.; Roberts, M.J.; Coughlin, G.D.; Lavin, M.F.; et al. Design and Clinical Verification of Surface-Enhanced Raman Spectroscopy Diagnostic Technology for Individual Cancer Risk Prediction. ACS Nano 2018, 12, 8362–8371. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Koo, K.M.; Wang, Y.; Trau, M. Engineering State-of-the-Art Plasmonic Nanomaterials for SERS-Based Clinical Liquid Biopsy Applications. Adv. Sci. 2019, 6, 1900730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Jiang, J. Embedding Raman Tags between Au Nanostar@Nanoshell for Multiplex Immunosensing. Small 2016, 12, 4980–4985. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.W.; So, P.T.; Dasari, R.R.; Lim, D.K. High resolution live cell Raman imaging using subcellular organelle-targeting SERS-sensitive gold nanoparticles with highly narrow intra-nanogap. Nano Lett. 2015, 15, 1766–1772. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, A.; Tian, L.; Tadepalli, S.; Liu, K.K.; Fei, M.; Farrell, M.E.; Pellegrino, P.M.; Singamaneni, S. Plasmonic nanorattles with intrinsic electromagnetic hot-spots for surface enhanced Raman scattering. Small 2014, 10, 4287–4292. [Google Scholar] [CrossRef]
- Gandra, N.; Singamaneni, S. Bilayered Raman-intense gold nanostructures with hidden tags (BRIGHTs) for high-resolution bioimaging. Adv. Mater. 2013, 25, 1022–1027. [Google Scholar] [CrossRef]
- Sun, L.; Sung, K.-B.; Dentinger, C.; Lutz, B.; Nguyen, L.; Zhang, J.; Qin, H.; Yamakawa, M.; Cao, M.; Lu, Y.; et al. Composite organic-inorganic nanoparticles as Raman labels for tissue analysis. Nano Lett. 2007, 7, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.K.; Jeon, K.S.; Hwang, J.H.; Kim, H.; Kwon, S.; Suh, Y.D.; Nam, J.M. Highly uniform and reproducible surface-enhanced Raman scattering from DNA-tailorable nanoparticles with 1-nm interior gap. Nat. Nanotech. 2011, 6, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.W.; Lim, D.K.; Kim, G.H.; Suh, Y.D.; Nam, J.M. Thiolated DNA-based chemistry and control in the structure and optical properties of plasmonic nanoparticles with ultrasmall interior nanogap. J. Am. Chem. Soc. 2014, 136, 14052–14059. [Google Scholar] [CrossRef]
- Bao, Z.; Zhang, Y.; Tan, Z.; Yin, X.; Di, W.; Ye, J. Gap-enhanced Raman tags for high-contrast sentinel lymph node imaging. Biomaterials 2018, 163, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qiu, Y.; Lin, L.; Gu, H.; Xiao, Z.; Ye, J. Ultraphotostable Mesoporous Silica-Coated Gap-Enhanced Raman Tags (GERTs) for High-Speed Bioimaging. ACS Appl. Mater. Interfaces 2017, 9, 3995–4005. [Google Scholar] [CrossRef]
- Nam, J.M.; Oh, J.W.; Lee, H.; Suh, Y.D. Plasmonic Nanogap-Enhanced Raman Scattering with Nanoparticles. Acc. Chem. Res. 2016, 49, 2746–2755. [Google Scholar] [CrossRef]
- Hu, C.; Shen, J.; Yan, J.; Zhong, J.; Qin, W.; Liu, R.; Aldalbahi, A.; Zuo, X.; Song, S.; Fan, C.; et al. Highly narrow nanogap-containing Au@Au core-shell SERS nanoparticles: Size-dependent Raman enhancement and applications in cancer cell imaging. Nanoscale 2016, 8, 2090–2096. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, Y.; He, J.; Thackray, B.D.; Ye, J. Ultrabright gap-enhanced Raman tags for high-speed bioimaging. Nat. Commun. 2019, 10, 3905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Shen, J.; Chen, S.; Wang, D.; Li, F.; Mathur, S.; Song, S.; Fan, C. Gold nanostructures encoded by non-fluorescent small molecules in polyA-mediated nanogaps as universal SERS nanotags for recognizing various bioactive molecules. Chem. Sci. 2014, 5, 4460–4466. [Google Scholar] [CrossRef]
- Shen, J.; Su, J.; Yan, J.; Zhao, B.; Wang, D.; Wang, S.; Li, K.; Liu, M.; He, Y.; Mathur, S.; et al. Bimetallic nano-mushrooms with DNA-mediated interior nanogaps for high-efficiency SERS signal amplification. Nano Res. 2014, 8, 731–742. [Google Scholar] [CrossRef]
- Kim, M.; Ko, S.M.; Kim, J.M.; Son, J.; Lee, C.; Rhim, W.K.; Nam, J.M. Dealloyed Intra-Nanogap Particles with Highly Robust, Quantifiable Surface-Enhanced Raman Scattering Signals for Biosensing and Bioimaging Applications. ACS Cent. Sci. 2018, 4, 277–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Gu, H.; Ye, J. Plasmonic multi-shell nanomatryoshka particles as highly tunable SERS tags with built-in reporters. Chem. Commun. 2015, 51, 17740–17743. [Google Scholar] [CrossRef]
- Madu, C.O.; Lu, Y. Novel diagnostic biomarkers for prostate cancer. J. Cancer 2010, 1, 150–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, R.A.; Schirra, H.J.; Catto, J.W.; Lavin, M.F.; Gardiner, R.A. Markers for detection of prostate cancer. Cancers 2010, 2, 1125–1154. [Google Scholar] [CrossRef] [Green Version]
- Cappuzzo, F.; Varella-Garcia, M.; Shigematsu, H.; Domenichini, I.; Bartolini, S.; Ceresoli, G.L.; Rossi, E.; Ludovini, V.; Gregorc, V.; Toschi, L. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor–positive non–small-cell lung cancer patients. J. Clin. Oncol. 2005, 23, 5007–5018. [Google Scholar] [CrossRef]
- Daniele, L.; Macrì, L.; Schena, M.; Dongiovanni, D.; Bonello, L.; Armando, E.; Ciuffreda, L.; Bertetto, O.; Bussolati, G.; Sapino, A. Predicting gefitinib responsiveness in lung cancer by fluorescence in situ hybridization/chromogenic in situ hybridization analysis of EGFR and HER2 in biopsy and cytology specimens. Mol. Cancer Ther. 2007, 6, 1223–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, F.; Varella-Garcia, M.; Cappuzzo, F. Predictive value of EGFR and HER2 overexpression in advanced non-small-cell lung cancer. Oncogene 2009, 28, S32–S37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.-K.; Kang, H.; Lee, H.; Jo, A.; Jeong, S.; Jeon, S.-J.; Kim, H.-I.; Lee, H.-Y.; Jeong, D.H.; Kim, J.-H.; et al. Single-Step and Rapid Growth of Silver Nanoshells as SERS-Active Nanostructures for Label-Free Detection of Pesticides. ACS Appl. Mater. Interfaces 2014, 6, 12541–12549. [Google Scholar] [CrossRef]
- Kang, H.; Yang, J.-K.; Noh, M.S.; Jo, A.; Jeong, S.; Lee, M.; Lee, S.; Chang, H.; Lee, H.; Jeon, S.-J.; et al. One-Step Synthesis of Silver Nanoshell with Bumps for Highly Sensitive Near-IR SERS Nanoprobes. J. Mater. Chem. B 2014, 2, 4415–4421. [Google Scholar] [CrossRef]
- Hong, S.Y.; Yeh, P.C.; Dadap, J.I.; Osgood, R.M. Jr. Interfacial dipole formation and surface-electron confinement in low-coverage self-assembled thiol layers: thiophenol and p-fluorothiophenol on Cu(111). ACS Nano 2012, 6, 10622–10631. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Deng, K.; Fichou, D.; Xie, S.S.; Nion, A.; Wang, C. STM imaging ortho- and para-fluorothiophenol self-assembled monolayers on Au(111). Langmuir 2009, 25, 5012–5017. [Google Scholar] [CrossRef]
- Jin, X.; Khlebtsov, B.N.; Khanadeev, V.A.; Khlebtsov, N.G.; Ye, J. Rational Design of Ultrabright SERS Probes with Embedded Reporters for Bioimaging and Photothermal Therapy. ACS Appl. Mater. Interfaces 2017, 9, 30387–30397. [Google Scholar] [CrossRef]
- Scholl, J.A.; García-Etxarri, A.; Koh, A.L.; Dionne, J.A. Observation of Quantum Tunneling between Two Plasmonic Nanoparticles. Nano Lett. 2013, 13, 564–569. [Google Scholar] [CrossRef]
- Lin, L.; Zapata, M.; Xiong, M.; Liu, Z.; Wang, S.; Xu, H.; Borisov, A.G.; Gu, H.; Nordlander, P.; Aizpurua, J.; et al. Nanooptics of Plasmonic Nanomatryoshkas: Shrinking the Size of a Core–Shell Junction to Subnanometer. Nano Lett. 2015, 15, 6419–6428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Wang, H.; Fang, Y.; Li, Z. Polarization State of Light Scattered from Quantum Plasmonic Dimer Antennas. ACS Nano 2016, 10, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Kang, H.; Ko, E.; Jun, B.-H.; Lee, H.-Y.; Lee, Y.-S.; Jeong, D.H. PSA Detection with Femtomolar Sensitivity and a Broad Dynamic Range Using SERS Nanoprobes and an Area-Scanning Method. ACS Sens. 2016, 1, 645–649. [Google Scholar] [CrossRef]
- Gao, Z.; Hou, L.; Xu, M.; Tang, D. Enhanced colorimetric immunoassay accompanying with enzyme cascade amplification strategy for ultrasensitive detection of low-abundance protein. Sci. Rep. 2014, 4, 3966. [Google Scholar] [CrossRef] [Green Version]
- Menju, T.; Hashimoto, S.; Hashimoto, A.; Otsuka, Y.; Handa, H.; Ogawa, E.; Toda, Y.; Wada, H.; Date, H.; Sabe, H. Engagement of overexpressed Her2 with GEP100 induces autonomous invasive activities and provides a biomarker for metastases of lung adenocarcinoma. PLoS ONE 2011, 6, e25301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, M.; Hirata, A.; Kometani, T.; Miyagawa, M.; Ueda, S.; Kinoshita, H.; Fujii, T.; Kuwano, M. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol. Cancer Ther. 2004, 3, 465–472. [Google Scholar]
- Kang, H.; Jeong, S.; Park, Y.; Yim, J.; Jun, B.-H.; Kyeong, S.; Yang, J.-K.; Kim, G.; Hong, S.; Lee, L.P.; et al. Near-infrared SERS nanoprobes with plasmonic Au/Ag hollow-shell assemblies for in vivo multiplex detection. Adv. Funct. Mater. 2013, 23, 3719–3727. [Google Scholar] [CrossRef]
- Mir-Simon, B.; Reche-Perez, I.; Guerrini, L.; Pazos-Perez, N.; Alvarez-Puebla, R.A. Universal One-Pot and Scalable Synthesis of SERS Encoded Nanoparticles. Chem. Mater. 2015, 27, 950–958. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, H.; Jeong, S.; Yang, J.-K.; Jo, A.; Lee, H.; Heo, E.H.; Jeong, D.H.; Jun, B.-H.; Chang, H.; Lee, Y.-S. Template-Assisted Plasmonic Nanogap Shells for Highly Enhanced Detection of Cancer Biomarkers. Int. J. Mol. Sci. 2021, 22, 1752. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22041752
Kang H, Jeong S, Yang J-K, Jo A, Lee H, Heo EH, Jeong DH, Jun B-H, Chang H, Lee Y-S. Template-Assisted Plasmonic Nanogap Shells for Highly Enhanced Detection of Cancer Biomarkers. International Journal of Molecular Sciences. 2021; 22(4):1752. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22041752
Chicago/Turabian StyleKang, Homan, Sinyoung Jeong, Jin-Kyoung Yang, Ahla Jo, Hyunmi Lee, Eun Hae Heo, Dae Hong Jeong, Bong-Hyun Jun, Hyejin Chang, and Yoon-Sik Lee. 2021. "Template-Assisted Plasmonic Nanogap Shells for Highly Enhanced Detection of Cancer Biomarkers" International Journal of Molecular Sciences 22, no. 4: 1752. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22041752