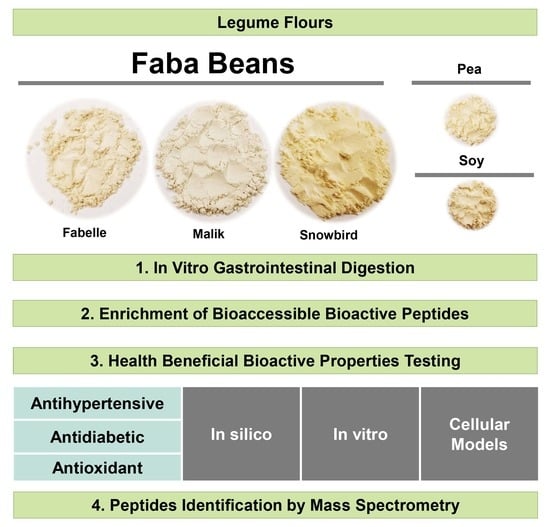
Health Beneficial Bioactivities of Faba Bean Gastrointestinal (In Vitro) Digestate in Comparison to Soybean and Pea
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of the In Vitro Digestion Model for Subsequent Cell Culture Experiments
2.2. In Silico Prediction of Bioactive Fragments Released during Gastrointestinal Digestion of Legume Proteins
2.3. Characterization of the 3 kDa Permeate of Legume Digestates
2.4. Bioactive Properties of the 3 kDa Permeate of Legume Digestates
2.4.1. Antioxidant and Chelating Activities
2.4.2. Antihypertensive Activity
2.4.3. Antidiabetic Activity
2.5. Peptides Fractionation and Sequencing
3. Materials and Methods
3.1. Materials
3.2. Sample Preparation
3.3. Adaptation of the In Vitro Gastrointestinal Digestion Procedure for Cell Culture
3.4. In Vitro Gastrointestinal Digestion of Legume Flours
3.5. In Silico Prediction of Bioactive Fragments Released during Gastrointestinal Digestion of Legume Proteins
3.6. Characterization of the 3 kDa Permeate of Legume Digestates
3.6.1. Molecular Weight Distribution of Peptides by Size Exclusion HPLC
3.6.2. Composition Analysis of the 3 kDa Permeate of Legume Digestates
3.7. In Vitro Antioxidant and Chelating Activities of the 3 kDa Permeate of Legume Digestates
3.7.1. 2,2-diphenyl-1-picrylhydrazyl (DPPH) Free Radical Scavenging Assay
3.7.2. 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) Scavenging Assay
3.7.3. Oxygen Radical Absorption Capacity (ORAC) Assay
3.7.4. Iron Chelating Activity
3.8. In Vitro Antihypertensive Activity (Angiotensin-Converting Enzyme Inhibition Activity)
3.9. In Vitro Antidiabetic Activity (Dipeptidyl Peptidase-IV Inhibition Assay)
3.10. Cell Culture
3.11. Cell Viability
3.12. Cellular Antioxidant Assay (CAA) in an Intestinal Cell Model
3.13. Dipeptidyl Peptidase-IV (DPP-IV) Inhibition in an Intestinal Cell Model
3.14. Faba Bean Peptides Fractionation and Sequencing by Mass Spectrometry
3.15. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Multari, S.; Stewart, D.; Russell, W.R. Potential of Fava Bean as Future Protein Supply to Partially Replace Meat Intake in the Human Diet. Compr. Rev. Food Sci. Food Saf. 2015, 14, 511–522. [Google Scholar] [CrossRef]
- FAOSTAT. FAOSTAT Database. Available online: http://www.fao.org/faostat/en/#home (accessed on 18 July 2022).
- Raikos, V.; Neacsu, M.; Russell, W.; Duthie, G. Comparative study of the functional properties of lupin, green pea, fava bean, hemp, and buckwheat flours as affected by pH. Food Sci. Nutr. 2014, 2, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Millar, K.A.; Gallagher, E.; Burke, R.; McCarthy, S.; Barry-Ryan, C. Proximate composition and anti-nutritional factors of fava-bean (Vicia faba), green-pea and yellow-pea (Pisum sativum) flour. J. Food Compos. Anal. 2019, 82, 103233. [Google Scholar] [CrossRef]
- Hou, W.; Zhang, X.; Yan, Q.; Li, P.; Sha, W.; Tian, Y.; Liu, Y. Linkage map of a gene controlling zero tannins (zt-1) In faba bean (Vicia faba L.) with SSR and ISSR markers. Agronomy 2018, 8, 80. [Google Scholar] [CrossRef]
- Khazaei, H.; Purves, R.W.; Hughes, J.; Link, W.; O’Sullivan, D.M.; Schulman, A.H.; Björnsdotter, E.; Geu-Flores, F.; Nadzieja, M.; Andersen, S.U.; et al. Eliminating vicine and convicine, the main anti-nutritional factors restricting faba bean usage. Trends Food Sci. Technol. 2019, 91, 549–556. [Google Scholar] [CrossRef]
- Gullón, P.; Gullón, B.; Tavaria, F.; Vasconcelos, M.; Gomes, A.M. In vitro fermentation of lupin seeds (Lupinus albus) and broad beans (Vicia faba): Dynamic modulation of the intestinal microbiota and metabolomic output. Food Funct. 2015, 6, 3316–3322. [Google Scholar] [CrossRef]
- Karataş, S.Ç.; Günay, D.; Sayar, S. In vitro evaluation of whole faba bean and its seed coat as a potential source of functional food components. Food Chem. 2017, 230, 182–188. [Google Scholar] [CrossRef]
- Rabey, J.M.; Vered, Y.; Shabtai, H.; Graff, E.; Korczyn, A.D. Improvement of parkinsonian features correlate with high plasma levodopa values after broad bean (Vicia faba) consumption. J. Neurol. Neurosurg. Psychiatry 1992, 55, 725–727. [Google Scholar] [CrossRef]
- Abdel-Sattar, E.; Mahrous, E.A.; Thabet, M.M.; Elnaggar, D.M.Y.; Youssef, A.M.; Elhawary, R.; Zaitone, S.A.; Celia Rodríguez, P.; Segura-Carretero, A.; Mekky, R.H. Methanolic extracts of a selected Egyptian Vicia faba cultivar mitigate the oxidative/inflammatory burden and afford neuroprotection in a mouse model of Parkinson’s disease. Inflammopharmacology 2021, 29, 221–235. [Google Scholar] [CrossRef]
- Johnson, J.B.; Skylas, D.J.; Mani, J.S.; Xiang, J.; Walsh, K.B.; Naiker, M. Phenolic Profiles of Ten Australian Faba Bean Varieties. Molecules 2021, 26, 4642. [Google Scholar] [CrossRef]
- Boudjou, S.; Oomah, B.D.; Zaidi, F.; Hosseinian, F. Phenolics content and antioxidant and anti-inflammatory activities of legume fractions. Food Chem. 2013, 138, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Belović, M.; Mastilović, J.; Torbica, A.; Tomić, J.R.; Stanić, D.R.; Džinić, N. Potential of bioactive proteins and peptides for prevention and treatment of mass non-communicable diseases. Food Feed. Res. 2011, 38, 51–61. [Google Scholar]
- Sánchez, A.; Vázquez, A. Bioactive peptides: A review. Food Qual. Saf. 2017, 1, 29–46. [Google Scholar] [CrossRef]
- León-Espinosa, E.B.; Sánchez-Chino, X.; Garduño-Siciliano, L.; Álvarez-González, R.I.; Dávila-Ortiz, G.; Madrigal-Bujaidar, E.; Téllez-Medina, D.I.; Jiménez-Martínez, C. Hypocholesterolemic and Anticarcinogenic Effect of Vicia faba Protein Hydrolyzates. Nutr. Cancer 2016, 68, 856–864. [Google Scholar] [CrossRef]
- Zambrowicz, A.; Pokora, M.; Setner, B.; Dąbrowska, A.; Szołtysik, M.; Babij, K.; Szewczuk, Z.; Trziszka, T.; Lubec, G.; Chrzanowska, J. Multifunctional peptides derived from an egg yolk protein hydrolysate: Isolation and characterization. Amino Acids 2015, 47, 369–380. [Google Scholar] [CrossRef]
- Felix, M.; Cermeño, M.; FitzGerald, R.J. Assessment of the microstructural characteristics and the in vitro bioactive properties of sunflower oil-based emulsions stabilized by fava bean (vicia faba) protein. Food Hydrocoll. 2019, 97, 105220. [Google Scholar] [CrossRef]
- Ashraf, J.; Awais, M.; Liu, L.; Khan, M.I.; Tong, L.-T.; Ma, Y.; Wang, L.; Zhou, X.; Zhou, S. Effect of thermal processing on cholesterol synthesis, solubilisation into micelles and antioxidant activities using peptides of Vigna angularis and Vicia faba. LWT 2020, 129, 109504. [Google Scholar] [CrossRef]
- Jakubczyk, A.; Karaś, M.; Złotek, U.; Szymanowska, U.; Baraniak, B.; Bochnak, J. Peptides obtained from fermented faba bean seeds (Vicia faba) as potential inhibitors of an enzyme involved in the pathogenesis of metabolic syndrome. LWT 2019, 105, 306–313. [Google Scholar] [CrossRef]
- Karkouch, I.; Tabbene, O.; Gharbi, D.; Ben Mlouka, M.A.; Elkahoui, S.; Rihouey, C.; Coquet, L.; Cosette, P.; Jouenne, T.; Limam, F. Antioxidant, antityrosinase and antibiofilm activities of synthesized peptides derived from Vicia faba protein hydrolysate: A powerful agents in cosmetic application. Ind. Crops Prod. 2017, 109, 310–319. [Google Scholar] [CrossRef]
- Dugardin, C.; Cudennec, B.; Tourret, M.; Caron, J.; Guérin-Deremaux, L.; Behra-Miellet, J.; Lefranc-Millot, C.; Ravallec, R. Explorative Screening of Bioactivities Generated by Plant-Based Proteins after In Vitro Static Gastrointestinal Digestion. Nutrients 2020, 12, 3746. [Google Scholar] [CrossRef]
- Narayanan, A.; Jones, L.H. Sulfonyl fluorides as privileged warheads in chemical biology. Chem. Sci. 2015, 6, 2650–2659. [Google Scholar] [CrossRef] [PubMed]
- Akter, S.; Addepalli, R.; Netzel, M.; Tinggi, U.; Fletcher, M.; Sultanbawa, Y.; Osborne, S. In vitro Bioaccessibility and Intestinal Absorption of Selected Bioactive Compounds in Terminalia ferdinandiana. Front. Nutr. 2021, 8, 818195. [Google Scholar] [CrossRef] [PubMed]
- Minekus, M.; Marie, A.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in-vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Egger, L.; Ménard, O.; Delgado-Andrade, C.; Alvito, P.; Assunção, R.; Balance, S.; Barberá, R.; Brodkorb, A.; Cattenoz, T.; Clemente, A.; et al. The harmonized INFOGEST in vitro digestion method: From knowledge to action. Food Res. Int. 2016, 88, 217–225. [Google Scholar] [CrossRef]
- Dziuba, M.; Dziuba, B. In silico analysis of bioactive peptides. In Bioactive Proteins and Peptides as Functional Foods and Nutraceuticals; Blackwell: Oxford, UK, 2010; pp. 325–340. [Google Scholar]
- Peach, M.J. Renin-angiotensin system: Biochemistry and mechanisms of action. Physiol. Rev. 1977, 57, 313–370. [Google Scholar] [CrossRef]
- Bischoff, H. The mechanism of alpha-glucosidase inhibition in the management of diabetes. Clin. Investig. Med. Med. Clin. Exp. 1995, 18, 303–311. [Google Scholar]
- Kumar, P.; Reithofer, V.; Reisinger, M.; Wallner, S.; Pavkov-Keller, T.; Macheroux, P.; Gruber, K. Substrate complexes of human dipeptidyl peptidase III reveal the mechanism of enzyme inhibition. Sci. Rep. 2016, 6, 23787. [Google Scholar] [CrossRef]
- Pang, X.; Shimizu, A.; Kurita, S.; Zankov, D.P.; Takeuchi, K.; Yasuda-Yamahara, M.; Kume, S.; Ishida, T.; Ogita, H. Novel Therapeutic Role for Dipeptidyl Peptidase III in the Treatment of Hypertension. Hypertension 2016, 68, 630–641. [Google Scholar] [CrossRef]
- Acquah, C.; Chan, Y.W.; Pan, S.; Agyei, D.; Udenigwe, C.C. Structure-informed separation of bioactive peptides. J. Food Biochem. 2019, 43, e12765. [Google Scholar] [CrossRef]
- Singh, B.; Singh, J.P.; Kaur, A.; Singh, N. Phenolic composition and antioxidant potential of grain legume seeds: A review. Food Res. Int. 2017, 101, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pechan, T.; Chang, S.K.C. Antioxidant and angiotensin-I converting enzyme inhibitory activities of phenolic extracts and fractions derived from three phenolic-rich legume varieties. J. Funct. Foods 2018, 42, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Al Shukor, N.; Van Camp, J.; Gonzales, G.B.; Staljanssens, D.; Struijs, K.; Zotti, M.J.; Raes, K.; Smagghe, G. Angiotensin-Converting Enzyme Inhibitory Effects by Plant Phenolic Compounds: A Study of Structure Activity Relationships. J. Agric. Food Chem. 2013, 61, 11832–11839. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, L.-j.; Zhu, J.-j.; Xu, B.; Li, R. Effect of soybean oligosaccharides on blood lipid, glucose levels and antioxidant enzymes activity in high fat rats. Food Chem. 2010, 119, 1633–1636. [Google Scholar] [CrossRef]
- Endringer, D.C.; Oliveira, O.V.; Braga, F.C. In vitro and in silico inhibition of angiotensin-converting enzyme by carbohydrates and cyclitols. Chem. Pap. 2014, 68, 37–45. [Google Scholar] [CrossRef]
- Martínez-Villaluenga, C.; Frías, J. Production and bioactivity of oligosaccharides in plant foods. In Food Oligosaccharides; John Wiley & Sons: Hoboken, NJ, USA, 2014; pp. 35–54. [Google Scholar] [CrossRef]
- Kedare, S.B.; Singh, R.P. Genesis and development of DPPH method of antioxidant assay. J. Food Sci. Technol. 2011, 48, 412–422. [Google Scholar] [CrossRef]
- Moniruzzaman, M.; Khalil, D.; Sulaiman, S.A.; Gan, S. Advances in the analytical methods for determining the antioxidant properties of honey. Afr. J. Tradit. Complement. Altern. Med. 2012, 9, 36–42. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free. Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Watanabe, J.; Oki, T.; Takebayashi, J.; Yamasaki, K.; Takano-Ishikawa, Y.; Hino, A.; Yasui, A. Method Validation by Interlaboratory Studies of Improved Hydrophilic Oxygen Radical Absorbance Capacity Methods for the Determination of Antioxidant Capacities of Antioxidant Solutions and Food Extracts. Anal. Sci. 2012, 28, 159. [Google Scholar] [CrossRef]
- Liang, N.; Kitts, D.D. Antioxidant property of coffee components: Assessment of methods that define mechanisms of action. Molecules 2014, 19, 19180–19208. [Google Scholar] [CrossRef]
- Zhong, Y.; Shahidi, F. 12—Methods for the assessment of antioxidant activity in foods. In Handbook of Antioxidants for Food Preservation; Shahidi, F., Ed.; Woodhead Publishing: Sawston, UK, 2015; pp. 287–333. [Google Scholar] [CrossRef]
- Gulcin, İ. Antioxidants and antioxidant methods: An updated overview. Arch. Toxicol. 2020, 94, 651–715. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, S.; Nie, S.; Yu, Q.; Xie, M. Reviews on Mechanisms of In Vitro Antioxidant Activity of Polysaccharides. Oxidative Med. Cell. Longev. 2016, 2016, 5692852. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.S.; Brizola, V.R.A.; Granato, D. High-throughput assay comparison and standardization for metal chelating capacity screening: A proposal and application. Food Chem. 2017, 214, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Parya Samaei, S.; Ghorbani, M.; Tagliazucchi, D.; Martini, S.; Gotti, R.; Themelis, T.; Tesini, F.; Gianotti, A.; Gallina Toschi, T.; Babini, E. Functional, nutritional, antioxidant, sensory properties and comparative peptidomic profile of faba bean (Vicia faba, L.) seed protein hydrolysates and fortified apple juice. Food Chem. 2020, 330, 127120. [Google Scholar] [CrossRef]
- Kellett, M.E.; Greenspan, P.; Pegg, R.B. Modification of the cellular antioxidant activity (CAA) assay to study phenolic antioxidants in a Caco-2 cell line. Food Chem. 2018, 244, 359–363. [Google Scholar] [CrossRef]
- Wolfe, K.L.; Liu, R.H. Cellular antioxidant activity (CAA) assay for assessing antioxidants, foods, and dietary supplements. J. Agric. Food Chem. 2007, 55, 8896–8907. [Google Scholar] [CrossRef]
- Wan, H.; Liu, D.; Yu, X.; Sun, H.; Li, Y. A Caco-2 cell-based quantitative antioxidant activity assay for antioxidants. Food Chem. 2015, 175, 601–608. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, H.; Liu, R.; Mats, L.; Zhu, H.; Pauls, K.P.; Deng, Z.; Tsao, R.J.J.o.F.F. Antioxidant and anti-inflammatory polyphenols and peptides of common bean (Phaseolus vulga L.) milk and yogurt in Caco-2 and HT-29 cell models. J. Funct. Foods 2019, 53, 125–135. [Google Scholar] [CrossRef]
- Torres-Fuentes, C.; Contreras, M.d.M.; Recio, I.; Alaiz, M.; Vioque, J. Identification and characterization of antioxidant peptides from chickpea protein hydrolysates. Food Chem. 2015, 180, 194–202. [Google Scholar] [CrossRef]
- Zhang, X.; Noisa, P.; Yongsawatdigul, J. Chemical and Cellular Antioxidant Activities of In Vitro Digesta of Tilapia Protein and Its Hydrolysates. Foods 2020, 9, 833. [Google Scholar] [CrossRef]
- Chaieb, N.; González, J.L.; López-Mesas, M.; Bouslama, M.; Valiente, M. Polyphenols content and antioxidant capacity of thirteen faba bean (Vicia faba L.) genotypes cultivated in Tunisia. Food Res. Int. 2011, 44, 970–977. [Google Scholar] [CrossRef]
- Caron, J.; Domenger, D.; Dhulster, P.; Ravallec, R.; Cudennec, B. Using Caco-2 cells as novel identification tool for food-derived DPP-IV inhibitors. Food Res. Int. 2017, 92, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, H.; Chen, S.; Luo, Y. Evaluating the effects of IADHFL on inhibiting DPP-IV activity and expression in Caco-2 cells and contributing to the amount of insulin released from INS-1 cells in vitro. Food Funct. 2018, 9, 2240–2250. [Google Scholar] [CrossRef] [PubMed]
- Lammi, C.; Bollati, C.; Ferruzza, S.; Ranaldi, G.; Sambuy, Y.; Arnoldi, A. Soybean-and lupin-derived peptides inhibit DPP-IV activity on in situ human intestinal Caco-2 cells and ex vivo human serum. Nutrients 2018, 10, 1082. [Google Scholar] [CrossRef] [PubMed]
- Aiello, G.; Li, Y.; Boschin, G.; Bollati, C.; Arnoldi, A.; Lammi, C. Chemical and biological characterization of spirulina protein hydrolysates: Focus on ACE and DPP-IV activities modulation. J. Funct. Foods 2019, 63, 103592. [Google Scholar] [CrossRef]
- Mamone, G.; Picariello, G.; Ramondo, A.; Nicolai, M.A.; Ferranti, P. Production, digestibility and allergenicity of hemp (Cannabis sativa L.) protein isolates. Food Res. Int. 2019, 115, 562–571. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Comparison of the susceptibility of porcine and human dipeptidyl-peptidase IV to inhibition by protein-derived peptides. Peptides 2015, 69, 19–25. [Google Scholar] [CrossRef]
- Alghamdi, S.S. Chemical composition of faba bean (Vicia faba L.) genotypes under various water regimes. Pak. J. Nutr. 2009, 8, 477–482. [Google Scholar] [CrossRef]
- Manzanares, P.; Gandía, M.; Garrigues, S.; Marcos, J.F. Improving Health-Promoting Effects of Food-Derived Bioactive Peptides through Rational Design and Oral Delivery Strategies. Nutrients 2019, 11, 2545. [Google Scholar] [CrossRef]
- Karami, Z.; Akbari-Adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- Lopez-Barrios, L.; Gutierrez-Uribe, J.A.; Serna-Saldivar, S.O. Bioactive peptides and hydrolysates from pulses and their potential use as functional ingredients. J. Food Sci. 2014, 79, R273–R283. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.L. Antioxidant peptides. In Bioactive Proteins and Peptides as Functional Foods and Nutraceuticals; Blackwell: Oxford, UK, 2010; pp. 29–42. [Google Scholar]
- Di Stefano, E.; Tsopmo, A.; Oliviero, T.; Fogliano, V.; Udenigwe, C.C. Bioprocessing of common pulses changed seed microstructures, and improved dipeptidyl peptidase-IV and α-glucosidase inhibitory activities. Sci. Rep. 2019, 9, 15308. [Google Scholar] [CrossRef] [PubMed]
- Loizzo, M.R.; Bonesi, M.; Leporini, M.; Falco, T.; Sicari, V.; Tundis, R. Chemical Profile and In Vitro Bioactivity of Vicia faba Beans and Pods. Proceedings 2021, 70, 45. [Google Scholar]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed]
- Dziuba, J.; Iwaniak, A.; Minkiewicz, P. Computer-aided characteristics of proteins as potential precursors of bioactive peptides. Polimery 2003, 48, 50–53. [Google Scholar] [CrossRef]
- L’Hocine, L.; Boye, J.I.; Arcand, Y. Composition and Functional Properties of Soy Protein Isolates Prepared Using Alternative Defatting and Extraction Procedures. J. Food Sci. 2006, 71, C137–C145. [Google Scholar] [CrossRef]
- Ma, Z.; Boye, J.I.; Simpson, B.K.; Prasher, S.O.; Monpetit, D.; Malcolmson, L. Thermal processing effects on the functional properties and microstructure of lentil, chickpea, and pea flours. Food Res. Int. 2011, 44, 2534–2544. [Google Scholar] [CrossRef]
- Hausch, F.; Shan, L.; Santiago, N.A.; Gray, G.M.; Khosla, C. Intestinal digestive resistance of immunodominant gliadin peptides. Am. J. Physiol. Liver Physiol. 2002, 283, G996–G1003. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Determination of the degree of hydrolysis of food protein hydrolysates by trinitrobenzenesulfonic acid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef]
- Mason, E. Nutritional and Bioactive Properties of Glabrous Canaryseed (Phalaris canariensis, L.) Proteins. Master’s Thesis, McGill University, Montreal, QC, Canada, 2019. [Google Scholar]
- Achouri, A.; Boye, J.I.; Belanger, D.; Chiron, T.; Yaylayan, V.A.; Yeboah, F.K. Functional and molecular properties of calcium precipitated soy glycinin and the effect of glycation with κ-carrageenan. Food Res. Int. 2010, 43, 1494–1504. [Google Scholar] [CrossRef]
- Mason, E.; L’Hocine, L.; Achouri, A.; Pitre, M.; Karboune, S. Health Promoting Bioactive Properties of Novel Hairless Canary Seed Flour after In Vitro Gastrointestinal Digestion. Foods 2020, 9, 932. [Google Scholar] [CrossRef] [PubMed]
- AOAC International (Ed.) Official Methods of Analysis of AOAC International; AOAC International: Arlington, TX, USA, 1995. [Google Scholar]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. [14] Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1999; Volume 299, pp. 152–178. [Google Scholar]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Orona-Tamayo, D.; Valverde, M.E.; Nieto-Rendón, B.; Paredes-López, O. Inhibitory activity of chia (Salvia hispanica L.) protein fractions against angiotensin I-converting enzyme and antioxidant capacity. LWT—Food Sci. Technol. 2015, 64, 236–242. [Google Scholar] [CrossRef]
- Tomer, D.P.; McLeman, L.D.; Ohmine, S.; Scherer, P.M.; Murray, B.K.; O’Neill, K.L. Comparison of the Total Oxyradical Scavenging Capacity and Oxygen Radical Absorbance Capacity Antioxidant Assays. J. Med. Food 2007, 10, 337–344. [Google Scholar] [CrossRef]
- Barbana, C.; Boye, J.I. Angiotensin I-converting enzyme inhibitory properties of lentil protein hydrolysates: Determination of the kinetics of inhibition. Food Chem. 2011, 127, 94–101. [Google Scholar] [CrossRef]
- Lammi, C.; Aiello, G.; Vistoli, G.; Zanoni, C.; Arnoldi, A.; Sambuy, Y.; Ferruzza, S.; Ranaldi, G. A multidisciplinary investigation on the bioavailability and activity of peptides from lupin protein. J. Funct. Foods 2016, 24, 297–306. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
- Zaky, A.A.; Simal-Gandara, J.; Eun, J.-B.; Shim, J.-H.; Abd El-Aty, A.M. Bioactivities, Applications, Safety, and Health Benefits of Bioactive Peptides from Food and By-Products: A Review. Front. Nutr. 2022, 8, 4646–4658. [Google Scholar] [CrossRef]












| Factor | p-Value 1 |
|---|---|
| Bile salt | |
| Bile salt concentration | 0.285 |
| Peptide concentration | <0.0001 |
| Bile salt concentration × Peptide concentration | 0.774 |
| Intestinal enzyme | |
| Intestinal enzyme used | <0.0001 |
| Peptide concentration | <0.0001 |
| Intestinal enzyme used × Peptide concentration | <0.0001 |
| Protease inhibitor | |
| Protease inhibitor concentration | <0.0001 |
| Peptide concentration | <0.0001 |
| Protease inhibitor concentration × Peptide concentration | 0.004 |
| % Proteins | Total Polyphenol (mg. Gallic Acid Equivalent/g) | Total Carbohydrates (g Glucose Equivalent/100 g) | |
|---|---|---|---|
| (Dry Base) | |||
| Fabelle | 34.37 ± 0.22 b | 4.81 ± 0.19 a | 43.5 ± 1.6 b |
| Malik | 36.11 ± 0.47 c | 4.97 ± 0.25 a | 44.0 ± 2.0 b |
| Snowbird | 38.42 ± 0.27 d | 4.80 ± 0.31 a | 45.2 ± 0.5 b |
| Amarillo | 32.87 ± 0.17 a | 4.43 ± 0.17 a | 46.7 ± 1.5 b |
| AAC-26-15 | 54.49 ± 0.54 e | 4.81 ± 0.51 a | 16.6 ± 0.8 a |
| ABTS 1 (% Scavenging) | Iron Chelating 2 (% Chelating) | ORAC (µmol Trolox eq/mg Peptides) | |
|---|---|---|---|
| F1 | 41.5 ± 10.4 c | 67.6 ± 2.7 b | 0.7 ± 0.5 b |
| F2 | 98.9 ± 0.8 a | 54.1 ± 1.8 c | 3.2 ± 0.6 a |
| F3 | 71.4 ± 15.3 b | 88.5 ± 1.9 a | 3.1 ± 0.1 a |
| Peptide Sequence | Fraction | Observed Mass (Da) | Calculated Mass (Da) | ppm 1 | Precursor Protein | Protein Accession Number | Fragment Location |
|---|---|---|---|---|---|---|---|
| N2YDEGSEPR | F1 | 1066.421 | 1066.420 | 0.43 | Convicilin | B0BCL8 | 29–37 |
| PVNRPGEPQ | F1 | 992.507 | 992.504 | 2.60 | Vicilin | I0B569 | 152–160 |
| LDNIN2ALEPDH | F1 | 1250.578 | 1250.578 | −0.19 | Legumin B | P05190.1 | 35–45 |
| TETWNPNHPE | F1 | 1223.522 | 1223.521 | 1.21 | Legumin B | P05190.1 | 52–61 |
| TETWNPNHPEL | F1 | 1336.606 | 1336.605 | 0.88 | Legumin B | P05190.1 | 52–62 |
| EEEDEDEPR | F1 | 1146.436 | 1146.431 | 3.64 | Legumin | Q43673 | 327–335 |
| KEEEDEDEPR | F1 | 1274.530 | 1274.526 | 3.06 | Legumin | Q43673 | 326–335 |
| VIPTEPPH | F1 | 888.470 | 888.471 | −0.70 | Tonoplast intrinsic protein 32 | A0A024NRI7 | 155–162 |
| VIPTEPPHA | F1 | 959.508 | 959.508 | −0.13 | Tonoplast intrinsic protein 32 | A0A024NRI7 | 155–163 |
| VVIPTEPPHA | F1 | 1058.577 | 1058.576 | 0.55 | Tonoplast intrinsic protein 32 | A0A024NRI7 | 154–163 |
| VVIPTEPPH | F1 and F2 | 987.540 | 987.539 | 1.07 | Tonoplast intrinsic protein 32 | A0A024NRI7 | 154–162 |
| Peptide Sequence | Fraction | Potential Bioactivity 1 | Bioactive Fragments | A 2 | B 3 (μM−1) |
|---|---|---|---|---|---|
| NYDEGSEPR | F1 | ACE inhibitor | PR, GS, EG, NY | 0.44 | 0.03 |
| Stimulating 5 | SE | 0.11 | . | ||
| DPP-IV inhibitor | EP, EG, NY, YD | 0.44 | . | ||
| DPP-III inhibitor | PR | 0.11 | . | ||
| PVNRPGEPQ | F1 | Anti-amnestic | PG | 0.11 | . |
| ACE inhibitor | GEP, RP, GE, PG, PQ | 0.56 | 0.36 | ||
| Antithrombotic | PG | 0.11 | . | ||
| Regulating 4 | PG | 0.11 | . | ||
| DPP-IV inhibitor | RP, EP, GE, NR, PG, PQ, PV, VN | 0.89 | 4.96 | ||
| DPP-III inhibitor | GE | 0.11 | . | ||
| Renin inhibitor | NR | 0.11 | . | ||
| LDNINALEPDH | F1 | ACE inhibitor | ALEP | 0.09 | 1.44 × 10−5 |
| DPP-IV inhibitor | EP, AL, DN, IN, NA | 0.45 | 1.03 × 10−4 | ||
| TETWNPNHPE | F1 | ACE inhibitor | TE, HP | 0.2 | . |
| Antioxidant | TW | 0.1 | . | ||
| Alpha-glucosidase inhibitor | PE | 0.1 | 3.99 × 10−6 | ||
| DPP-IV inhibitor | HP, NP, WN, ET, NH, PN, TE, TW | 0.8 | 3.35 × 10−3 | ||
| DPP-III inhibitor | HP, PE | 0.2 | . | ||
| TETWNPNHPEL | F1 | ACE inhibitor | TE, HP | 0.18 | . |
| Antioxidant | EL, PEL, TW, TETWNPNHPEL | 0.36 | . | ||
| Alpha-glucosidase inhibitor | PE | 0.09 | 3.63 × 10−6 | ||
| DPP-IV inhibitor | HP, NP, WN, ET, NH, PN, TE, TW | 0.73 | 3.04 × 10−3 | ||
| DPP-III inhibitor | HP,PE | 0.18 | . | ||
| EEEDEDEPR | F1 | ACE inhibitor | PR | 0.11 | 0.03 |
| Stimulating 5 | EEE, EE | 0.33 | . | ||
| DPP-IV inhibitor | EP | 0.11 | . | ||
| DPP-III inhibitor | PR | 0.11 | . | ||
| KEEEDEDEPR | F1 | ACE inhibitor | PR, KE | 0.2 | 0.02 |
| Stimulating 5 | EEE, EE | 0.3 | . | ||
| DPP-IV inhibitor | EP, KE | 0.2 | . | ||
| DPP-III inhibitor | PR | 0.1 | . | ||
| VIPTEPPH | F1 | ACE inhibitor | IP, TE, PT, PP, PH | 0.63 | 9.62 × 10−4 |
| Alpha-glucosidase inhibitor | PP | 0.13 | 6.93 × 10−6 | ||
| DPP-IV inhibitor | PP, IP, EP, PH, PT, TE, VI | 0.88 | 3.26 × 10−4 | ||
| VIPTEPPHA | F1 | ACE inhibitor | IP, TE, PT, PP, PH | 0.56 | 8.55 × 10−4 |
| Antioxidant | PHA | 0.11 | . | ||
| Alpha-glucosidase inhibitor | PP | 0.11 | 6.16 × 10−6 | ||
| DPP-IV inhibitor | PP, HA, IP, EP, PH, PT, TE, VI | 0.89 | 2.90 × 10−4 | ||
| VVIPTEPPHA | F1 | ACE inhibitor | IP, TE, PT, PP, PH | 0.5 | 7.69 × 10−4 |
| Antioxidant | PHA | 0.1 | . | ||
| Alpha-glucosidase inhibitor | PP | 0.1 | 5.55 × 10−6 | ||
| DPP-IV inhibitor | PP, VV, HA, IP, EP, PH, PT, TE, VI | 0.9 | 2.61 × 10−4 | ||
| VVIPTEPPH | F1 and F2 | ACE inhibitor | IP, TE, PT, PP, PH | 0.56 | 8.55 × 10−4 |
| Alpha-glucosidase inhibitor | PP | 0.11 | 6.16 × 10−6 | ||
| DPP-IV inhibitor | PP, VV, IP, EP, PH, PT, TE, VI | 0.89 | 2.90 × 10−4 |
| Faba Bean (Vicia faba) | |||
| Proteins | Gene | Accession Number | |
| Legumin (11S) | Legumin B | LEB4 | P05190 |
| LEB2 | P16078 | ||
| LEB6 | P16079 | ||
| LEB7 | P16080 | ||
| Legumin A | A1 | Q03971 | |
| A2 | Q99304 | ||
| Vicilin (7S) | Vicilin | . | P08438 |
| Convicilin | . | B0BCL8 | |
| . | B0BCL7 | ||
| Pea (Pisum sativum) | |||
| Proteins | Gene | Accession Number | |
| Legumin (11S) | Legumin B | LEGJ | P05692 |
| LEGK | P05693 | ||
| LEGB | P14594 | ||
| Legumin A | A | P02857 | |
| A2 | P15838 | ||
| Vicilin (7S) | Vicilin | . | P13918 |
| Convicilin | CVA | P13915 | |
| CVB | P13919 | ||
| Soy (Glycine max) | |||
| Proteins | Gene | Accession Number | |
| Glycinin (11S) | Glycinin-G1 | GY1 | P04776 |
| Glycinin-G2 | GY2 | P04405 | |
| Glycinin-G3 | GY3 | P11828 | |
| Glycinin-G4 | GY4 | P02858 | |
| Glycinin-G5 | GY5 | P04347 | |
| β-Conglycinin (7S) | β-Conglycinin-α’ | CG-1 | P11827 |
| β-Conglycinin-α | CG-3 | P0DO16 | |
| β-Conglycinin β | CG-4 | P25974 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martineau-Côté, D.; Achouri, A.; Wanasundara, J.; Karboune, S.; L’Hocine, L. Health Beneficial Bioactivities of Faba Bean Gastrointestinal (In Vitro) Digestate in Comparison to Soybean and Pea. Int. J. Mol. Sci. 2022, 23, 9210. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23169210
Martineau-Côté D, Achouri A, Wanasundara J, Karboune S, L’Hocine L. Health Beneficial Bioactivities of Faba Bean Gastrointestinal (In Vitro) Digestate in Comparison to Soybean and Pea. International Journal of Molecular Sciences. 2022; 23(16):9210. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23169210
Chicago/Turabian StyleMartineau-Côté, Delphine, Allaoua Achouri, Janitha Wanasundara, Salwa Karboune, and Lamia L’Hocine. 2022. "Health Beneficial Bioactivities of Faba Bean Gastrointestinal (In Vitro) Digestate in Comparison to Soybean and Pea" International Journal of Molecular Sciences 23, no. 16: 9210. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms23169210