Identification of an Optimal TLR8 Ligand by Alternating the Position of 2′-O-Ribose Methylation
Abstract
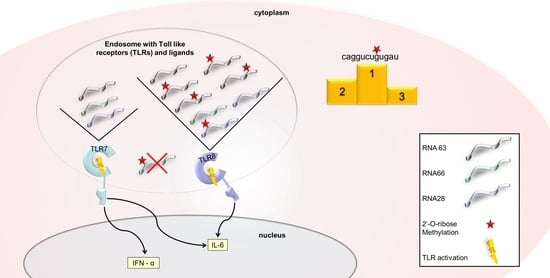
:1. Introduction
2. Results
2.1. 2′-O-Ribose Methylation Prevents TLR7 Activation Independent of the Position
 ). In contrast, the analysis of RNA63M2 and RNA63M4 digests showed an altered pattern in this region resembling a single band or two co-migrating bands. Furthermore, digested RNA63M1 and RNA63M3 showed two additional small bands (compared with a single smaller band in digested RNA63M2 and RNA63M4), which is marked with a line (
). In contrast, the analysis of RNA63M2 and RNA63M4 digests showed an altered pattern in this region resembling a single band or two co-migrating bands. Furthermore, digested RNA63M1 and RNA63M3 showed two additional small bands (compared with a single smaller band in digested RNA63M2 and RNA63M4), which is marked with a line ( ). Overall, these patterns suggest a difference in ORN fragments created by RNase T2 digestion.
). Overall, these patterns suggest a difference in ORN fragments created by RNase T2 digestion.2.2. Impact of 2′-O-Ribose Methylation on TLR7 Activation by Naturally Methylated and Unmethylated Sequences
2.3. Activation of TLR8 by 2′-O-Ribose Methylated ORNs
3. Discussion
4. Materials and Methods
4.1. Kits and Reagents
4.2. Cells
4.3. Cell Stimulation
4.4. Enzyme-Linked Immunosorbent Assay (ELISA)
4.5. Genetic Complementation Assay
4.6. Mice
4.7. RNase T2 Digestion Assay
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
 ) and a line (
) and a line ( ). Three independent replicates of RNase T2 digestion are shown.
). Three independent replicates of RNase T2 digestion are shown.
 ) and a line (
) and a line ( ). Three independent replicates of RNase T2 digestion are shown.
). Three independent replicates of RNase T2 digestion are shown.
References
- Chan, Y.K.; Gack, M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016, 14, 360–373. [Google Scholar] [CrossRef]
- Murphy, K.; Weaver, C. Janeway′s Immunobiology; Garland Science: New York, NY, USA, 2017. [Google Scholar]
- Lind, N.A.; Rael, V.E.; Pestal, K.; Liu, B.; Barton, G.M. Regulation of the nucleic acid-sensing Toll-like receptors. Nat. Rev. Immunol. 2022, 22, 224–235. [Google Scholar] [CrossRef]
- Cervantes, J.L.; Weinerman, B.; Basole, C.; Salazar, J.C. TLR8: The forgotten relative revindicated. Cell Mol. Immunol. 2012, 9, 434–438. [Google Scholar] [CrossRef]
- Bender, A.T.; Tzvetkov, E.; Pereira, A.; Wu, Y.; Kasar, S.; Przetak, M.M.; Vlach, J.; Niewold, T.B.; Jensen, M.A.; Okitsu, S.L. TLR7 and TLR8 Differentially Activate the IRF and NF-κB Pathways in Specific Cell Types to Promote Inflammation. Immunohorizons 2020, 4, 93–107. [Google Scholar] [CrossRef]
- Hornung, V.; Guenthner-Biller, M.; Bourquin, C.; Ablasser, A.; Schlee, M.; Uematsu, S.; Noronha, A.; Manoharan, M.; Akira, S.; de Fougerolles, A.; et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005, 11, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Poeck, H.; Anz, D.; Berger, M.; Schlee, M.; Kim, S.; Bourquin, C.; Goutagny, N.; Jiang, Z.; Fitzgerald, K.A.; et al. Selection of molecular structure and delivery of RNA oligonucleotides to activate TLR7 versus TLR8 and to induce high amounts of IL-12p70 in primary human monocytes. J. Immunol. 2009, 182, 6824–6833. [Google Scholar] [CrossRef] [PubMed]
- de Marcken, M.; Dhaliwal, K.; Danielsen, A.C.; Gautron, A.S.; Dominguez-Villar, M. TLR7 and TLR8 activate distinct pathways in monocytes during RNA virus infection. Sci. Signal. 2019, 12, eaaw1347. [Google Scholar] [CrossRef]
- Bartok, E.; Hartmann, G. Immune Sensing Mechanisms that Discriminate Self from Altered Self and Foreign Nucleic Acids. Immunity 2020, 53, 54–77. [Google Scholar] [CrossRef]
- Hartmann, G. Nucleic Acid Immunity. Adv. Immunol. 2017, 133, 121–169. [Google Scholar] [CrossRef]
- Barchet, W.; Wimmenauer, V.; Schlee, M.; Hartmann, G. Accessing the therapeutic potential of immunostimulatory nucleic acids. Curr. Opin. Immunol. 2008, 20, 389–395. [Google Scholar] [CrossRef]
- Hemmi, H.; Kaisho, T.; Takeuchi, O.; Sato, S.; Sanjo, H.; Hoshino, K.; Horiuchi, T.; Tomizawa, H.; Takeda, K.; Akira, S. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat. Immunol. 2002, 3, 196–200. [Google Scholar] [CrossRef]
- Jurk, M.; Heil, F.; Vollmer, J.; Schetter, C.; Krieg, A.M.; Wagner, H.; Lipford, G.; Bauer, S. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat. Immunol. 2002, 3, 499. [Google Scholar] [CrossRef]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar] [CrossRef]
- Krüger, A.; Oldenburg, M.; Chebrolu, C.; Beisser, D.; Kolter, J.; Sigmund, A.M.; Steinmann, J.; Schäfer, S.; Hochrein, H.; Rahmann, S.; et al. Human TLR8 senses UR/URR motifs in bacterial and mitochondrial RNA. EMBO Rep. 2015, 16, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Forsbach, A.; Nemorin, J.G.; Montino, C.; Müller, C.; Samulowitz, U.; Vicari, A.P.; Jurk, M.; Mutwiri, G.K.; Krieg, A.M.; Lipford, G.B.; et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 2008, 180, 3729–3738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ohto, U.; Shibata, T.; Taoka, M.; Yamauchi, Y.; Sato, R.; Shukla, N.M.; David, S.A.; Isobe, T.; Miyake, K.; et al. Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands. Cell Rep. 2018, 25, 3371–3381.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ohto, U.; Shibata, T.; Krayukhina, E.; Taoka, M.; Yamauchi, Y.; Tanji, H.; Isobe, T.; Uchiyama, S.; Miyake, K.; et al. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity 2016, 45, 737–748. [Google Scholar] [CrossRef]
- Shibata, T.; Ohto, U.; Nomura, S.; Kibata, K.; Motoi, Y.; Zhang, Y.; Murakami, Y.; Fukui, R.; Ishimoto, T.; Sano, S.; et al. Guanosine and its modified derivatives are endogenous ligands for TLR7. Int. Immunol. 2016, 28, 211–222. [Google Scholar] [CrossRef]
- Ostendorf, T.; Zillinger, T.; Andryka, K.; Schlee-Guimaraes, T.M.; Schmitz, S.; Marx, S.; Bayrak, K.; Linke, R.; Salgert, S.; Wegner, J.; et al. Immune Sensing of Synthetic, Bacterial, and Protozoan RNA by Toll-like Receptor 8 Requires Coordinated Processing by RNase T2 and RNase 2. Immunity 2020, 52, 591–605.e6. [Google Scholar] [CrossRef] [PubMed]
- Greulich, W.; Wagner, M.; Gaidt, M.M.; Stafford, C.; Cheng, Y.; Linder, A.; Carell, T.; Hornung, V. TLR8 Is a Sensor of RNase T2 Degradation Products. Cell 2019, 179, 1264–1275.e13. [Google Scholar] [CrossRef]
- Tanji, H.; Ohto, U.; Shibata, T.; Taoka, M.; Yamauchi, Y.; Isobe, T.; Miyake, K.; Shimizu, T. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 2015, 22, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Damase, T.R.; Sukhovershin, R.; Boada, C.; Taraballi, F.; Pettigrew, R.I.; Cooke, J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 628137. [Google Scholar] [CrossRef]
- Auderset, F.; Belnoue, E.; Mastelic-Gavillet, B.; Lambert, P.H.; Siegrist, C.A. A TLR7/8 Agonist-Including DOEPC-Based Cationic Liposome Formulation Mediates Its Adjuvanticity Through the Sustained Recruitment of Highly Activated Monocytes in a Type I IFN-Independent but NF-κB-Dependent Manner. Front. Immunol. 2020, 11, 580974. [Google Scholar] [CrossRef] [PubMed]
- Mackman, R.L.; Mish, M.; Chin, G.; Perry, J.K.; Appleby, T.; Aktoudianakis, V.; Metobo, S.; Pyun, P.; Niu, C.; Daffis, S.; et al. Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B. J. Med. Chem. 2020, 63, 10188–10203. [Google Scholar] [CrossRef] [PubMed]
- Amin, O.E.; Colbeck, E.J.; Daffis, S.; Khan, S.; Ramakrishnan, D.; Pattabiraman, D.; Chu, R.; Micolochick Steuer, H.; Lehar, S.; Peiser, L.; et al. Therapeutic Potential of TLR8 Agonist GS-9688 (Selgantolimod) in Chronic Hepatitis B: Remodeling of Antiviral and Regulatory Mediators. Hepatology 2021, 74, 55–71. [Google Scholar] [CrossRef]
- Freund, I.; Buhl, D.K.; Boutin, S.; Kotter, A.; Pichot, F.; Marchand, V.; Vierbuchen, T.; Heine, H.; Motorin, Y.; Helm, M.; et al. 2′-O-methylation within prokaryotic and eukaryotic tRNA inhibits innate immune activation by endosomal Toll-like receptors but does not affect recognition of whole organisms. RNA 2019, 25, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Freund, I.; Eigenbrod, T.; Helm, M.; Dalpke, A.H. RNA Modifications Modulate Activation of Innate Toll-Like Receptors. Genes 2019, 10, 92. [Google Scholar] [CrossRef]
- Schmitt, F.C.F.; Freund, I.; Weigand, M.A.; Helm, M.; Dalpke, A.H.; Eigenbrod, T. Identification of an optimized 2′-O-methylated trinucleotide RNA motif inhibiting Toll-like receptors 7 and 8. RNA 2017, 23, 1344–1351. [Google Scholar] [CrossRef]
- Hamm, S.; Latz, E.; Hangel, D.; Muller, T.; Yu, P.; Golenbock, D.; Sparwasser, T.; Wagner, H.; Bauer, S. Alternating 2′-O-ribose methylation is a universal approach for generating non-stimulatory siRNA by acting as TLR7 antagonist. Immunobiology 2009, 215, 559–569. [Google Scholar] [CrossRef]
- Jockel, S.; Nees, G.; Sommer, R.; Zhao, Y.; Cherkasov, D.; Hori, H.; Ehm, G.; Schnare, M.; Nain, M.; Kaufmann, A.; et al. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 2012, 209, 235–241. [Google Scholar] [CrossRef]
- Jung, S.; von Thülen, T.; Laukemper, V.; Pigisch, S.; Hangel, D.; Wagner, H.; Kaufmann, A.; Bauer, S. A single naturally occurring 2′-O-methylation converts a TLR7- and TLR8-activating RNA into a TLR8-specific ligand. PLoS ONE 2015, 10, e0120498. [Google Scholar] [CrossRef]
- Gehrig, S.; Eberle, M.E.; Botschen, F.; Rimbach, K.; Eberle, F.; Eigenbrod, T.; Kaiser, S.; Holmes, W.M.; Erdmann, V.A.; Sprinzl, M.; et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 2012, 209, 225–233. [Google Scholar] [CrossRef]
- Lee, J.; Chuang, T.H.; Redecke, V.; She, L.; Pitha, P.M.; Carson, D.A.; Raz, E.; Cottam, H.B. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: Activation of Toll-like receptor 7. Proc. Natl. Acad. Sci. USA 2003, 100, 6646–6651. [Google Scholar] [CrossRef]
- Piekna-Przybylska, D.; Decatur, W.A.; Fournier, M.J. The 3D rRNA modification maps database: With interactive tools for ribosome analysis. Nucleic Acids Res. 2008, 36, D178–D183. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef]
- Sarvestani, S.T.; Williams, B.R.; Gantier, M.P. Human Toll-like receptor 8 can be cool too: Implications for foreign RNA sensing. J. Interferon Cytokine Res. 2012, 32, 350–361. [Google Scholar] [CrossRef]
- Krug, A.; Rothenfusser, S.; Hornung, V.; Jahrsdörfer, B.; Blackwell, S.; Ballas, Z.K.; Endres, S.; Krieg, A.M.; Hartmann, G. Identification of CpG oligonucleotide sequences with high induction of IFN-alpha/beta in plasmacytoid dendritic cells. Eur. J. Immunol. 2001, 31, 2154–2163. [Google Scholar] [CrossRef]
- Lu, H.; Dietsch, G.N.; Matthews, M.A.; Yang, Y.; Ghanekar, S.; Inokuma, M.; Suni, M.; Maino, V.C.; Henderson, K.E.; Howbert, J.J.; et al. VTX-2337 is a novel TLR8 agonist that activates NK cells and augments ADCC. Clin. Cancer Res. 2012, 18, 499–509. [Google Scholar] [CrossRef]
- Czauderna, F.; Fechtner, M.; Dames, S.; Aygün, H.; Klippel, A.; Pronk, G.J.; Giese, K.; Kaufmann, J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003, 31, 2705–2716. [Google Scholar] [CrossRef]
- Obermann, H.L.; Lederbogen, I.I.; Steele, J.; Dorna, J.; Sander, L.E.; Engelhardt, K.; Bakowsky, U.; Kaufmann, A.; Bauer, S. RNA-Cholesterol Nanoparticles Function as Potent Immune Activators via TLR7 and TLR8. Front. Immunol. 2021, 12, 658895. [Google Scholar] [CrossRef]
- Lan, T.; Putta, M.R.; Wang, D.; Dai, M.; Yu, D.; Kandimalla, E.R.; Agrawal, S. Synthetic oligoribonucleotides-containing secondary structures act as agonists of Toll-like receptors 7 and 8. Biochem. Biophys. Res. Commun. 2009, 386, 443–448. [Google Scholar] [CrossRef]
- Spies, B.; Hochrein, H.; Vabulas, M.; Huster, K.; Busch, D.H.; Schmitz, F.; Heit, A.; Wagner, H. Vaccination with plasmid DNA activates dendritic cells via Toll-like receptor 9 (TLR9) but functions in TLR9-deficient mice. J. Immunol. 2003, 171, 5908–5912. [Google Scholar] [CrossRef] [Green Version]
 ) and a line (
) and a line ( ). (c,d) PBMCs were stimulated with RNA63 derivatives at a final concentration of 10 µg/mL: 1 µM CpG ODN 2216 or 5 µg/mL RNA40. Supernatants were harvested 20 h post-stimulation (h p. s.) and both (c) IFN-α and (d) IL-6 concentrations were measured in ELISA. Graphs depict six independent experiments with PBMCs obtained from six individual donors, each in biological duplicates (twelve measurements per data point, mean + S.D). Data were analyzed using paired t-tests. **** p < 0.0001, *** p < 0.001.
). (c,d) PBMCs were stimulated with RNA63 derivatives at a final concentration of 10 µg/mL: 1 µM CpG ODN 2216 or 5 µg/mL RNA40. Supernatants were harvested 20 h post-stimulation (h p. s.) and both (c) IFN-α and (d) IL-6 concentrations were measured in ELISA. Graphs depict six independent experiments with PBMCs obtained from six individual donors, each in biological duplicates (twelve measurements per data point, mean + S.D). Data were analyzed using paired t-tests. **** p < 0.0001, *** p < 0.001.
 ) and a line (
) and a line ( ). (c,d) PBMCs were stimulated with RNA63 derivatives at a final concentration of 10 µg/mL: 1 µM CpG ODN 2216 or 5 µg/mL RNA40. Supernatants were harvested 20 h post-stimulation (h p. s.) and both (c) IFN-α and (d) IL-6 concentrations were measured in ELISA. Graphs depict six independent experiments with PBMCs obtained from six individual donors, each in biological duplicates (twelve measurements per data point, mean + S.D). Data were analyzed using paired t-tests. **** p < 0.0001, *** p < 0.001.
). (c,d) PBMCs were stimulated with RNA63 derivatives at a final concentration of 10 µg/mL: 1 µM CpG ODN 2216 or 5 µg/mL RNA40. Supernatants were harvested 20 h post-stimulation (h p. s.) and both (c) IFN-α and (d) IL-6 concentrations were measured in ELISA. Graphs depict six independent experiments with PBMCs obtained from six individual donors, each in biological duplicates (twelve measurements per data point, mean + S.D). Data were analyzed using paired t-tests. **** p < 0.0001, *** p < 0.001.




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicolai, M.; Steinberg, J.; Obermann, H.-L.; Solis, F.V.; Bartok, E.; Bauer, S.; Jung, S. Identification of an Optimal TLR8 Ligand by Alternating the Position of 2′-O-Ribose Methylation. Int. J. Mol. Sci. 2022, 23, 11139. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms231911139
Nicolai M, Steinberg J, Obermann H-L, Solis FV, Bartok E, Bauer S, Jung S. Identification of an Optimal TLR8 Ligand by Alternating the Position of 2′-O-Ribose Methylation. International Journal of Molecular Sciences. 2022; 23(19):11139. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms231911139
Chicago/Turabian StyleNicolai, Marina, Julia Steinberg, Hannah-Lena Obermann, Francisco Venegas Solis, Eva Bartok, Stefan Bauer, and Stephanie Jung. 2022. "Identification of an Optimal TLR8 Ligand by Alternating the Position of 2′-O-Ribose Methylation" International Journal of Molecular Sciences 23, no. 19: 11139. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms231911139