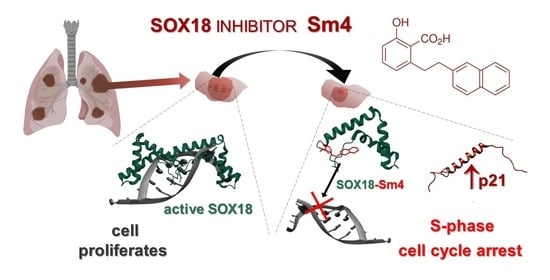
Targeting SOX18 Transcription Factor Activity by Small-Molecule Inhibitor Sm4 in Non-Small Lung Cancer Cell Lines
Abstract
:1. Introduction
2. Results
2.1. Sm4 Treatment Shows Comparable Cytotoxicity in NSCLC and IMR-90 Cell Lines
2.2. Disruption of SOX18 Activity Alters Transcript Levels of SOX7 and SOX17 but Not Protein Expression
2.3. Sm4-Mediated Cell Cycle Arrest Exhibits a Prominent S-phase Cell Accumulation in LXF-289 Cell Line
2.4. Differential Effects of Sm4 on Cyclin Expression in LXF-289 and SK-MES-1 Cell Lines
2.5. Sm4-Induced SOX18 Activity Inhibition Upregulated p21 Expression
2.6. SOX18 and p21 IHC Expression Levels Are Negatively Correlated in Lung Cancer Tissue Samples
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Experimental Design
4.3. Cell Culture
4.4. MTT Assay
4.5. Cell Cycle Assay
4.6. Gene Expression by qPCR
4.7. Western Blot Analysis
4.8. Immunohistochemistry (IHC)
4.9. Evaluation of IHC Reactions
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gubbay, J.; Collignon, J.; Koopman, P.; Capel, B.; Economou, A.; Münsterberg, A.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 1990, 346, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Francois, M.; Koopman, P.; Beltrame, M. SoxF genes: Key players in the development of the cardio-vascular system. Int. J. Biochem. Cell Biol. 2010, 42, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Downes, M.; Koopman, P. SOX18 and the transcriptional regulation of blood vessel development. Trends Cardiovasc. Med. 2001, 11, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.I.S.; Ma, A.C.H.; Fung, T.-K.; Leung, A.Y.H. Characterization of Sry-related HMG box group F genes in zebrafish hematopoiesis. Exp. Hematol. 2011, 39, 986–998.e5. [Google Scholar] [CrossRef]
- François, M.; Caprini, A.; Hosking, B.; Orsenigo, F.; Wilhelm, D.; Browne, C.; Paavonen, K.; Karnezis, T.; Shayan, R.; Downes, M.; et al. Sox18 induces development of the lymphatic vasculature in mice. Nature 2008, 456, 643–647. [Google Scholar] [CrossRef]
- Gross, C.M.; Aggarwal, S.; Kumar, S.; Tian, J.; Kasa, A.; Bogatcheva, N.; Datar, S.A.; Verin, A.D.; Fineman, J.R.; Black, S.M. Sox18 Preserves the Pulmonary Endothelial Barrier Under Conditions of Increased Shear Stress. J. Cell. Physiol. 2014, 229, 1802–1816. [Google Scholar] [CrossRef] [Green Version]
- Roudnicky, F.; Kim, B.K.; Lan, Y.; Schmucki, R.; Küppers, V.; Christensen, K.; Graf, M.; Patsch, C.; Burcin, M.; Meyer, C.A.; et al. Identification of a combination of transcription factors that synergistically increases endothelial cell barrier resistance. Sci. Rep. 2020, 10, 3886. [Google Scholar] [CrossRef] [Green Version]
- Darby, I.A.; Bisucci, T.; Raghoenath, S.; Olsson, J.; Muscat, G.E.; Koopman, P. Sox18 Is Transiently Expressed during Angiogenesis in Granulation Tissue of Skin Wounds with an Identical Expression Pattern to Flk-1 mRNA. Lab. Investig. 2001, 81, 937–943. [Google Scholar] [CrossRef] [Green Version]
- Saitoh, T.; Katoh, M. Expression of human SOX18 in normal tissues and tumors. Int. J. Mol. Med. 2002, 10, 339–344. [Google Scholar] [CrossRef]
- Matsui, T.; Kanai-Azuma, M.; Hara, K.; Matoba, S.; Hiramatsu, R.; Kawakami, H.; Kurohmaru, M.; Koopman, P.; Kanai, Y. Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. J. Cell Sci. 2006, 119, 3513–3526. [Google Scholar] [CrossRef] [Green Version]
- Grimm, D.; Bauer, J.; Wise, P.; Krüger, M.; Simonsen, U.; Wehland, M.; Infanger, M.; Corydon, T.J. The role of SOX family members in solid tumours and metastasis. Semin. Cancer Biol. 2019, 67, 122–153. [Google Scholar] [CrossRef] [PubMed]
- Olbromski, M.; Dzięgiel, P. Role of the SOX18 protein in neoplastic processes. Oncol. Lett. 2018, 16, 1383–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olbromski, M.; Podhorska-Okołów, M.; Dzięgiel, P. Role of SOX Protein Groups F and H in Lung Cancer Progression. Cancers 2020, 12, 3235. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Lei, B.; Huang, Q. SOX18 Affects Cell Viability, Migration, Invasiveness, and Apoptosis in Hepatocellular Carcinoma (HCC) Cells by Participating in Epithelial-to-Mesenchymal Transition (EMT) Progression and Adenosine Monophosphate Activated Protein Kinase (AMPK)/Mammalian Target of Rapamycin (mTOR). Med. Sci. Monit. 2019, 25, 6244–6254. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Li, Z.; Zhao, D.; Hui, B.; Zheng, Z. SOX18 enhances the proliferation and migration of airway smooth muscle cells induced by tumor necrosis factor-α via the regulation of Notch1 signaling. Int. Immunopharmacol. 2021, 96, 107746. [Google Scholar] [CrossRef]
- Geng, Q.; Deng, H.; Fu, J.; Cui, F. SOX18 exerts tumor-suppressive functions in papillary thyroid carcinoma through inhibition of Wnt/β-catenin signaling. Exp. Cell Res. 2020, 396, 112249. [Google Scholar] [CrossRef]
- Wu, D.; Jiang, C.; Zheng, J.-J.; Luo, D.-S.; Ma, J.; Que, H.-F.; Li, C.; Ma, C.; Wang, H.-Y.; Wang, W.; et al. Bioinformatics analysis of SOXF family genes reveals potential regulatory mechanism and diagnostic value in cancers. Ann. Transl. Med. 2022, 10, 701. [Google Scholar] [CrossRef]
- Hou, L.; Srivastava, Y.; Jauch, R. Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 2017, 63, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Fontaine, F.R.; Goodall, S.; Prokop, J.W.; Howard, C.B.; Moustaqil, M.; Kumble, S.; Rasicci, D.T.; Osborne, G.W.; Gambin, Y.; Sierecki, E.; et al. Functional domain analysis of SOX18 transcription factor using a single-chain variable fragment-based approach. mAbs 2018, 10, 596–606. [Google Scholar] [CrossRef] [Green Version]
- Moustaqil, M.; Fontaine, F.; Overman, J.; McCann, A.; Bailey, T.L.; Soto, P.R.; Bhumkar, A.; Giles, N.; Hunter, D.J.B.; Gambin, Y.; et al. Homodimerization regulates an endothelial specific signature of the SOX18 transcription factor. Nucleic Acids Res. 2018, 46, 11381–11395. [Google Scholar] [CrossRef] [Green Version]
- Jethon, A.; Pula, B.; Olbromski, M.; Werynska, B.; Muszczynska-Bernhard, B.; Witkiewicz, W.; Dziegiel, P.; Podhorska-Okolow, M. Prognostic significance of SOX18 expression in non-small cell lung cancer. Int. J. Oncol. 2015, 46, 123–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pula, B.; Olbromski, M.; Wojnar, A.; Gomulkiewicz, A.; Witkiewicz, W.; Ugorski, M.; Dziegiel, P.; Podhorska-Okolow, M. Impact of SOX18 expression in cancer cells and vessels on the outcome of invasive ductal breast carcinoma. Cell. Oncol. 2013, 36, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Q.; Wu, P.; Xiang, W.; Xie, D.; Wang, N.; Deng, M.; Cao, K.; Zeng, H.; Xu, Z.; et al. SLC12A5 interacts and enhances SOX18 activity to promote bladder urothelial carcinoma progression via upregulating MMP7. Cancer Sci. 2020, 111, 2349–2360. [Google Scholar] [CrossRef] [PubMed]
- Ornat, M.; Kobierzycki, C.; Grzegrzolka, J.; Pula, B.; Zamirska, A.; Bieniek, A.; Szepietowski, J.C.; Dziegiel, P.; Podhorska-Okolow, M. SOX18 Expression in Non-melanoma Skin Cancer. Anticancer Res. 2016, 36, 2379–2383. [Google Scholar]
- Yin, H.; Qin, C.; Wang, Q.; Du, Y.; Xu, T. The role of SOX18 in bladder cancer and its underlying mechanism in mediating cellular functions. Life Sci. 2019, 232, 116614. [Google Scholar] [CrossRef]
- Yin, H.; Sheng, Z.; Zhang, X.; Du, Y.; Qin, C.; Liu, H.; Dun, Y.; Wang, Q.; Jin, C.; Zhao, Y.; et al. Overexpression of SOX18 promotes prostate cancer progression via the regulation of TCF1, c-Myc, cyclin D1 and MMP-7. Oncol. Rep. 2017, 37, 1045–1051. [Google Scholar] [CrossRef] [Green Version]
- Miao, Z.; Deng, X.; Shuai, P.; Zeng, J. Upregulation of SOX18 in colorectal cancer cells promotes proliferation and correlates with colorectal cancer risk. OncoTargets Ther. 2018, 11, 8481–8490. [Google Scholar] [CrossRef] [Green Version]
- Petrovic, I.; Milivojevic, M.; Popovic, J.; Schwirtlich, M.; Rankovic, B.; Stevanovic, M. SOX18 Is a Novel Target Gene of Hedgehog Signaling in Cervical Carcinoma Cell Lines. PLoS ONE 2015, 10, e0143591. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Wang, Y.; Zhang, D.; Yu, X.; Leng, X. MiR-7-5p Functions as a Tumor Suppressor by Targeting SOX18 in Pancreatic Ductal Adenocarcinoma. Biochem. Biophys. Res. Commun. 2018, 497, 963–970. [Google Scholar] [CrossRef]
- Chen, J.; Dang, Y.; Feng, W.; Qiao, C.; Liu, D.; Zhang, T.; Wang, Y.; Tian, D.; Fan, D.; Nie, Y.; et al. SOX18 promotes gastric cancer metastasis through transactivating MCAM and CCL7. Oncogene 2020, 39, 5536–5552. [Google Scholar] [CrossRef]
- Yin, H.; Qin, C.; Wang, Q.; Du, Y.; Dai, X.; Tang, X.; Zhang, X.; Li, Q.; Liu, S.; Xu, T. Transcription Factor SOX18 Promotes Clear Cell Renal Cell Carcinoma Progression and Alleviates Cabozantinib-Mediated Inhibitory Effects. Mol. Cancer Ther. 2019, 18, 2433–2445. [Google Scholar] [CrossRef] [Green Version]
- Lawrenson, K.; Fonseca, M.A.; Liu, A.Y.; Dezem, F.S.; Lee, J.M.; Lin, X.; Corona, R.I.; Abbasi, F.; Vavra, K.C.; Dinh, H.Q.; et al. A Study of High-Grade Serous Ovarian Cancer Origins Implicates the SOX18 Transcription Factor in Tumor Development. Cell Rep. 2019, 29, 3726–3735.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Liu, J.; Wang, J.; Zhang, F. SOX18 knockdown suppresses the proliferation and metastasis, and induces the apoptosis of osteosarcoma cells. Mol. Med. Rep. 2015, 13, 497–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, N.; Hahn, C.N.; Poh, A.; Dong, C.; Wilhelm, D.; Olsson, J.; Muscat, G.E.O.; Parsons, P.; Gamble, J.R.; Koopman, P. Effect of Disrupted SOX18 Transcription Factor Function on Tumor Growth, Vascularization, and Endothelial Development. J. Natl. Cancer Inst. 2006, 98, 1060–1067. [Google Scholar] [CrossRef]
- Zhu, D.; Yang, D.; Li, X.; Feng, F. Heterogeneous expression and biological function of SOX18 in osteosaroma. J. Cell. Biochem. 2017, 119, 4184–4192. [Google Scholar] [CrossRef]
- Darnell, J.E. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2002, 2, 740–749. [Google Scholar] [CrossRef]
- Fontaine, F.; Overman, J.; Moustaqil, M.; Mamidyala, S.; Salim, A.; Narasimhan, K.; Prokoph, N.; Robertson, A.A.; Lua, L.; Alexandrov, K.; et al. Small-Molecule Inhibitors of the SOX18 Transcription Factor. Cell Chem. Biol. 2017, 24, 346–359. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Guo, H.; Zhang, D.; Yu, X.; Leng, X.; Li, S.; Zhu, W. Overexpression of SOX18 correlates with accelerated cell growth and poor prognosis in human pancreatic ductal adenocarcinoma. Biochem. Biophys. Res. Commun. 2016, 479, 510–516. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, Y.; Wang, S.; Chen, F.; Gu, Y. Suppression of SOX18 by siRNA inhibits cell growth and invasion of breast cancer cells. Oncol. Rep. 2016, 35, 3721–3727. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Wei, Z.; Jia, H.; Zhao, W.; Yang, G.; Zhao, H. Knockdown of SOX18 inhibits the proliferation, migration and invasion of hepatocellular carcinoma cells. Oncol. Rep. 2015, 34, 1121–1128. [Google Scholar] [CrossRef] [Green Version]
- Overman, J.; Fontaine, F.; Moustaqil, M.; Mittal, D.; Sierecki, E.; Sacilotto, N.; Zuegg, J.; Robertson, A.A.B.; Holmes, K.; Salim, A.A.; et al. Pharmacological targeting of the transcription factor SOX18 delays breast cancer in mice. eLife 2017, 6, e21221. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rodak, O.; Peris-Díaz, M.D.; Olbromski, M.; Podhorska-Okołów, M.; Dzięgiel, P. Current Landscape of Non-Small Cell Lung Cancer: Epidemiology, Histological Classification, Targeted Therapies, and Immunotherapy. Cancers 2021, 13, 4705. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Heigener, D.F.; Mok, T.; Soria, J.-C.; Rabe, K.F. Management of non-small-cell lung cancer: Recent developments. Lancet 2013, 382, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Azhikina, T.; Kozlova, A.; Skvortsov, T.; Sverdlov, E. Heterogeneity and degree of TIMP4, GATA4, SOX18, and EGFL7 gene promoter methylation in non–small cell lung cancer and surrounding tissues. Cancer Genet. 2011, 204, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.; Strunnikova, M.; Schagdarsurengin, U.; Rastetter, M.; Papritz, M.; Hattenhorst, U.E.; Hofmann, H.-S.; Silber, R.-E.; Burdach, S.; Hansen, G. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur. J. Cancer 2005, 41, 1223–1236. [Google Scholar] [CrossRef]
- Olbromski, M.; Grzegrzolka, J.; Jankowska-Konsur, A.; Witkiewicz, W.; Podhorska-Okolow, M.; Dziegiel, P. MicroRNAs modulate the expression of the SOX18 transcript in lung squamous cell carcinoma. Oncol. Rep. 2016, 36, 2884–2892. [Google Scholar] [CrossRef] [Green Version]
- Olbromski, M.; Rzechonek, A.; Grzegrzółka, J.; Glatzel-Plucinska, N.; Chachaj, A.; Werynska, B.; Podhorska-Okolow, M.; Dziegiel, P. Influence of miR-7a and miR-24-3p on the SOX18 transcript in lung adenocarcinoma. Oncol. Rep. 2017, 39, 201–208. [Google Scholar] [CrossRef] [Green Version]
- Cermenati, S.; Moleri, S.; Cimbro, S.; Corti, P.; Del Giacco, L.; Amodeo, R.; Dejana, E.; Koopman, P.; Cotelli, F.; Beltrame, M. Sox18 and Sox7 play redundant roles in vascular development. Blood 2008, 111, 2657–2666. [Google Scholar] [CrossRef] [Green Version]
- Tuohinto, K.; DiMaio, T.A.; Kiss, E.A.; Laakkonen, P.; Saharinen, P.; Karnezis, T.; Lagunoff, M.; Ojala, P.M. KSHV infection of endothelial precursor cells with lymphatic characteristics as a novel model for translational Kaposi’s sarcoma studies. PLoS Pathog. 2023, 19, e1010753. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Hara, K.; Kanai-Azuma, M.; Matsui, T.; Miura, Y.; Tsunekawa, N.; Kurohmaru, M.; Saijoh, Y.; Koopman, P.; Kanai, Y. Redundant roles of Sox17 and Sox18 in early cardiovascular development of mouse embryos. Biochem. Biophys. Res. Commun. 2007, 360, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.G.; Gandillet, A.; Pearson, S.; Lacaud, G.; Kouskoff, V. Contrasting effects of Sox17- and Sox18-sustained expression at the onset of blood specification. Blood 2010, 115, 3895–3898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Williams, J.; Smallwood, P.M.; Nathans, J. Sox7, Sox17, and Sox18 Cooperatively Regulate Vascular Development in the Mouse Retina. PLoS ONE 2015, 10, e0143650. [Google Scholar] [CrossRef] [Green Version]
- Stovall, D.B.; Cao, P.; Sui, G. SOX7: From a developmental regulator to an emerging tumor suppressor. Histol. Histopathol. 2013, 29, 439–445. [Google Scholar] [PubMed]
- Zhang, Y.; Huang, S.; Dong, W.; Li, L.; Feng, Y.; Pan, L.; Han, Z.; Wang, X.; Ren, G.; Su, D.; et al. SOX7, down-regulated in colorectal cancer, induces apoptosis and inhibits proliferation of colorectal cancer cells. Cancer Lett. 2009, 277, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Hayano, T.; Garg, M.; Yin, D.; Sudo, M.; Kawamata, N.; Shi, S.; Chien, W.; Ding, L.-W.; Leong, G.; Mori, S.; et al. SOX7 is down-regulated in lung cancer. J. Exp. Clin. Cancer Res. 2013, 32, 86. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Lv, Z.; He, G.; Wang, J.; Zhang, X.; Lu, G.; Ren, X.; Wang, F.; Zhu, X.; Ding, Y.; et al. The SOX17/miR-371-5p/SOX2 axis inhibits EMT, stem cell properties and metastasis in colorectal cancer. Oncotarget 2015, 6, 9099–9112. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Stovall, D.B.; Wan, M.; Zhang, Q.; Chou, J.W.; Li, D.; Sui, G. SOX7 Target Genes and Their Contribution to Its Tumor Suppressive Function. Int. J. Mol. Sci. 2018, 19, 1451. [Google Scholar] [CrossRef] [Green Version]
- Niculescu, A.B.; Chen, X.; Smeets, M.; Hengst, L.; Prives, C.; Reed, S.I. Effects of P21 Cip1/Waf1 at Both the G 1 /S and the G 2 /M Cell Cycle Transitions: PRb Is a Critical Determinant in Blocking DNA Replication and in Preventing Endoreduplication. Mol. Cell. Biol. 1998, 18, 629–643. [Google Scholar] [CrossRef] [Green Version]
- Dulić, V.; Stein, G.H.; Far, D.F.; Reed, S.I. Nuclear Accumulation of P21 Cip1 at the Onset of Mitosis: A Role at the G 2 /M-Phase Transition. Mol. Cell. Biol. 1998, 18, 546–557. [Google Scholar] [CrossRef] [Green Version]
- Maharjan, S.; Kwon, Y.S.; Lee, M.G.; Lee, K.S.; Nam, K.S. Cell Cycle Arrest-Mediated Cell Death by Morin in MDA-MB-231 Triple-Negative Breast Cancer Cells. Pharmacol. Rep. 2021, 73, 1315–1327. [Google Scholar] [CrossRef]
- Huang, B.; Mu, P.; Chen, X.; Tang, S.; Ye, W.; Zhu, W.; Deng, Y. Aflatoxin B1 Induces S Phase Arrest by Upregulating the Expression of P21 via MYC, PLK1 and PLD1. Biochem. Pharmacol. 2019, 166, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.K.; Dana, S.; Mandal, K.; Mukhopadhyay, P.; Mondal, N. Downregulation of C-Myc and P21 Expression and Induction of S Phase Arrest by Naphthalene Diimide Derivative in Gastric Adenocarcinoma Cells. Chem. Biol. Interact. 2019, 304, 106–123. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, L.; Wu, S.; Teraishi, F.; Davis, J.J.; Jacob, D.; Fang, B. Induction of S-Phase Arrest and P21 Overexpression by a Small Molecule 2[[3-(2,3-Dichlorophenoxy)Propyl] Amino]Ethanol in Correlation with Activation of ERK. Oncogene 2004, 23, 4984–4992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shishodia, S.; Sethi, G.; Ahn, K.S.; Aggarwal, B.B. Guggulsterone Inhibits Tumor Cell Proliferation, Induces S-Phase Arrest, and Promotes Apoptosis through Activation of c-Jun N-Terminal Kinase, Suppression of Akt Pathway, and Downregulation of Antiapoptotic Gene Products. Biochem. Pharmacol. 2007, 74, 118–130. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Chen, D.B.; Lin, C.; Cao, K.; Wan, Y.; Zhao, X.Y.; Nie, C.L.; Yuan, Z.; Wei, Y.Q. HPNAS-4 Inhibits Proliferation through S Phase Arrest and Apoptosis: Underlying Action Mechanism in Ovarian Cancer Cells. Apoptosis 2013, 18, 467–479. [Google Scholar] [CrossRef]
- Abbas, T.; Dutta, A. P21 in Cancer: Intricate Networks and Multiple Activities. Nat Rev Cancer 2009, 9, 400. [Google Scholar] [CrossRef]
- Harper, J.W.; Elledge, S.J.; Keyomarsi, K.; Dynlacht, B.; Tsai, L.H.; Zhang, P.; Dobrowolski, S.; Bai, C.; Connell-Crowley, L.; Swindell, E.; et al. Inhibition of Cyclin-Dependent Kinases by P21. Mol Biol Cell 1995, 6, 387–400. [Google Scholar] [CrossRef] [Green Version]
- Mansilla, S.F.; de la Vega, M.B.; Calzetta, N.L.; Siri, S.O.; Gottifredi, V. CDK-Independent and PCNA-Dependent Functions of P21 in DNA Replication. Genes 2020, 11, 593. [Google Scholar] [CrossRef]
- Duong, T.; Koltowska, K.; Pichol-Thievend, C.; Le Guen, L.; Fontaine, F.; Smith, K.A.; Truong, V.; Skoczylas, R.; Stacker, S.A.; Achen, M.G.; et al. VEGFD Regulates Blood Vascular Development by Modulating SOX18 Activity. Blood 2014, 123, 1102–1112. [Google Scholar] [CrossRef] [Green Version]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodak, O.; Mrozowska, M.; Rusak, A.; Gomułkiewicz, A.; Piotrowska, A.; Olbromski, M.; Podhorska-Okołów, M.; Ugorski, M.; Dzięgiel, P. Targeting SOX18 Transcription Factor Activity by Small-Molecule Inhibitor Sm4 in Non-Small Lung Cancer Cell Lines. Int. J. Mol. Sci. 2023, 24, 11316. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms241411316
Rodak O, Mrozowska M, Rusak A, Gomułkiewicz A, Piotrowska A, Olbromski M, Podhorska-Okołów M, Ugorski M, Dzięgiel P. Targeting SOX18 Transcription Factor Activity by Small-Molecule Inhibitor Sm4 in Non-Small Lung Cancer Cell Lines. International Journal of Molecular Sciences. 2023; 24(14):11316. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms241411316
Chicago/Turabian StyleRodak, Olga, Monika Mrozowska, Agnieszka Rusak, Agnieszka Gomułkiewicz, Aleksandra Piotrowska, Mateusz Olbromski, Marzenna Podhorska-Okołów, Maciej Ugorski, and Piotr Dzięgiel. 2023. "Targeting SOX18 Transcription Factor Activity by Small-Molecule Inhibitor Sm4 in Non-Small Lung Cancer Cell Lines" International Journal of Molecular Sciences 24, no. 14: 11316. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms241411316