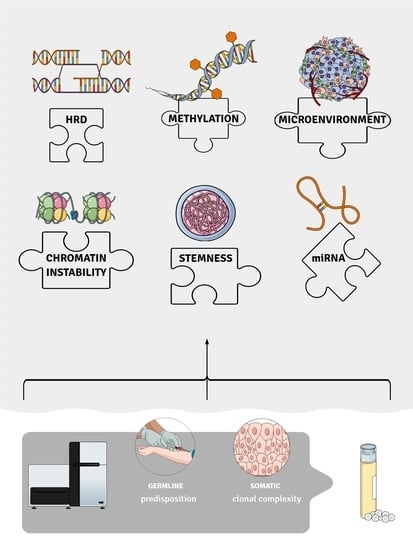
A Glance at Molecular Advances in Cancer Genetics: A Baffling Puzzle Still to Be Solved
Author Contributions
Funding
Conflicts of Interest
References
- Zuntini, R.; Bonora, E.; Pradella, L.M.; Amato, L.B.; Vidone, M.; De Fanti, S.; Catucci, I.; Cortesi, L.; Medici, V.; Ferrari, S.; et al. Detecting variants in the nbn gene while testing for hereditary breast cancer: What to do next? Int. J. Mol. Sci. 2021, 22, 5832. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Hirasawa, A. Homologous Recombination Deficiencies and Hereditary Tumors. Int. J. Mol. Sci. 2021, 23, 348. [Google Scholar] [CrossRef] [PubMed]
- Sahnane, N.; Carnevali, I.; Formenti, G.; Casarin, J.; Facchi, S.; Bombelli, R.; Di Lauro, E.; Memoli, D.; Salvati, A.; Rizzo, F.; et al. BRCA methylation testing identifies a subset of ovarian carcinomas without germline variants that can benefit from PARP inhibitor. Int. J. Mol. Sci. 2020, 21, 9708. [Google Scholar] [CrossRef] [PubMed]
- Wadowska, K.; Bil-Lula, I.; Trembecki, Ł.; Śliwińska-Mossoń, M. Genetic markers in lung cancer diagnosis: A review. Int. J. Mol. Sci. 2020, 21, 4569. [Google Scholar] [CrossRef]
- Urazbakhtin, S.; Smirnova, A.; Volakhava, A.; Zerkalenkova, E.; Salyutina, M.; Doubek, M.; Jelinkova, H.; Khudainazarova, N.; Volchkov, E.; Belyaeva, L.; et al. The Absence of Retroelement Activity Is Characteristic for Childhood Acute Leukemias and Adult Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2022, 23, 1756. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.; Park, S.J.; Lee, G.; Yoon, S.K.; Lee, M.; Lee, S.K. The gnaq t96s mutation affects cell signaling and enhances the oncogenic properties of hepatocellular carcinoma. Int. J. Mol. Sci. 2021, 22, 3284. [Google Scholar] [CrossRef] [PubMed]
- Dameri, M.; Ferrando, L.; Cirmena, G.; Vernieri, C.; Pruneri, G.; Ballestrero, A.; Zoppoli, G. Multi-gene testing overview with a clinical perspective in metastatic triple-negative breast cancer. Int. J. Mol. Sci. 2021, 22, 7154. [Google Scholar] [CrossRef] [PubMed]
- Bresadola, L.; Weber, D.; Ritzel, C.; Löwer, M.; Bukur, V.; Akilli-öztürk, Ö.; Becker, J.; Mehanna, H.; Schrörs, B.; Vascotto, F.; et al. Comprehensive genomic and transcriptomic analysis of three synchronous primary tumours and a recurrence from a head and neck cancer patient. Int. J. Mol. Sci. 2021, 22, 7583. [Google Scholar] [CrossRef] [PubMed]
- Rontauroli, S.; Carretta, C.; Parenti, S.; Bertesi, M.; Manfredini, R. Novel Molecular Insights into Leukemic Evolution of Myeloproliferative Neoplasms: A Single Cell Perspective. Int. J. Mol. Sci. 2022, 23, 15256. [Google Scholar] [CrossRef]
- Carretta, C.; Mallia, S.; Genovese, E.; Parenti, S.; Rontauroli, S.; Bianchi, E.; Fantini, S.; Sartini, S.; Tavernari, L.; Tagliafico, E.; et al. Genomic analysis of hematopoietic stem cell at the single-cell level: Optimization of cell fixation and whole genome amplification (WGA) protocol. Int. J. Mol. Sci. 2020, 21, 7366. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, L.; Grillo, F.; Albertelli, M.; Ghiorzo, P.; Bruno, W. Insights into mechanisms of tumorigenesis in neuroendocrine neoplasms. Int. J. Mol. Sci. 2021, 22, 10328. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghiorzo, P.; Bruno, W. A Glance at Molecular Advances in Cancer Genetics: A Baffling Puzzle Still to Be Solved. Int. J. Mol. Sci. 2023, 24, 1394. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms24021394
Ghiorzo P, Bruno W. A Glance at Molecular Advances in Cancer Genetics: A Baffling Puzzle Still to Be Solved. International Journal of Molecular Sciences. 2023; 24(2):1394. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms24021394
Chicago/Turabian StyleGhiorzo, Paola, and William Bruno. 2023. "A Glance at Molecular Advances in Cancer Genetics: A Baffling Puzzle Still to Be Solved" International Journal of Molecular Sciences 24, no. 2: 1394. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms24021394