Polymer-Degrading Enzymes of Pseudomonas chloroaphis PA23 Display Broad Substrate Preferences
Abstract
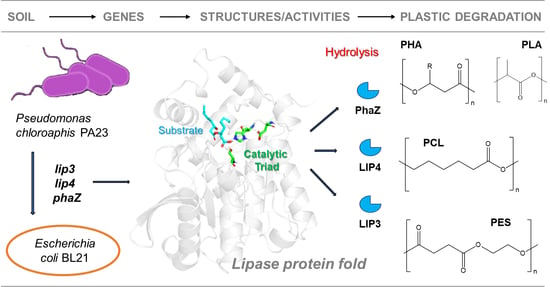
:1. Introduction
2. Results
2.1. Cloning, Expression and Purification of LIP3, LIP4 and PhaZ
2.2. Sequence Analyses and Homology Modeling of LIP3, LIP4, and PhaZ
2.3. Characterization of the Recombinant LIP3, LIP4, and PhaZ
2.4. Substrate Specificity of LIP3, LIP4, and PhaZ
2.5. Analysis of Degradation Products
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Plasmids
4.2. Chemicals, Media and Growth Conditions
4.3. DNA Manipulation and Plasmid Construction
4.4. Sequence Analysis and Homology Modeling
4.5. Expression and Purification of Recombinant LIP3, LIP4 and PhaZ
4.6. Qualitative and Quantitative Estimation of Depolymerase/Esterase Activity of LIP3, LIP4, and PhaZ
4.7. Biophysical and Biochemical Properties and Substrate Specificity of LIP3, LIP4 and PhaZ
4.8. Gel Permeation Chromatography
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wilkes, R.A.; Aristilde, L. Degradation and metabolism of synthetic plastics and associated products by Pseudomonas sp.: Capabilities and challenges. J. Appl. Microbiol. 2017, 123, 582–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernando, W.G.D.; Nakkeeran, S.; Zhang, Y.; Savchuk, S. Biological control of Sclerotinia sclerotiorum (Lib.) de Bary by Pseudomonas and Bacillus species on canola petals. Crop Prot. 2007, 26, 100–107. [Google Scholar] [CrossRef]
- Savchuk, S.C.; Fernando, D.W.G. Effect of timing of application and population dynamics on the degree of biological control of Sclerotinia sclerotiorum by bacterial antagonists. FEMS Microbiol. Ecol. 2004, 49, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Jendrossek, D.; Handrick, R. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 2002, 56, 403–432. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.; Kim, T.D.; Kim, K.K. Carboxylic ester hydrolases in bacteria: Active site, structure, function and application. Crystals 2019, 9, 597. [Google Scholar] [CrossRef] [Green Version]
- Reis, P.; Holmberg, K.; Watzke, H.; Leser, M.E.; Miller, R. Lipases at interfaces: A review. Adv. Colloid Interface Sci. 2009, 147, 237–250. [Google Scholar] [CrossRef]
- Fojan, P.; Jonson, P.H.; Petersen, M.T.N.; Petersen, S.B. What distinguishes an esterase from a lipase: A novel structural approach. Biochimie 2000, 82, 1033–1041. [Google Scholar] [CrossRef]
- Chandra, R.; Rustgi, R. Biodegradable polymers. Prog. Polym. Sci. 1998, 23, 1273–1335. [Google Scholar] [CrossRef]
- Okada, M. Chemical syntheses of biodegradable polymers. Prog. Polym. Sci. 2002, 27, 87–133. [Google Scholar] [CrossRef]
- Knoll, M.; Hamm, T.M.; Wagner, F.; Martinez, V.; Pleiss, J. The PHA depolymerase engineering database: A systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinf. 2009, 10, 89. [Google Scholar] [CrossRef] [Green Version]
- Abas, M.; Bahadur, A.; Ashraf, Z.; Iqbal, S.; Rajoka, M.S.R.; Rashid, S.G.; Jabeen, E.; Iqbal, Z.; Abbas, Q.; Bais, A.; et al. Designing novel anticancer sulfonamide based 2,5-disubstituted-1,3,4-thiadiazole derivatives as potential carbonic anhydrase inhibitor. J. Mol. Struct. 2021, 1246, 131145. [Google Scholar] [CrossRef]
- Hussain, R.; Shah, M.; Iqbal, S.; Rehman, W.; Khan, S.; Rasheed, L.; Naz, H.; Al-ghulikah, H.A.; Elkaeed, E.B.; Pashameah, R.A.; et al. Molecular iodine-promoted oxidative cyclization for the synthesis of 1,3,4-thiadiazole-fused-[1,2,4]-thiadiazole incorporating 1,4-benzodioxine moiety as potent inhibitors of α-amylase and α-glucosidase: In vitro and in silico study. Front. Chem. 2022, 10, 1023316. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, U.; Brzozowski, A.M.; Lawson, D.M.; Derewenda, Z.S. Catalysis at the interface: The anatomy of a conformational change in a triglyceride lipase. Biochemistry 1992, 11, 1532–1541. [Google Scholar] [CrossRef] [PubMed]
- Longhi, S.; Czjzek, M.; Lamzin, V.; Nicolas, A.; Cambillau, C. Atomic resolution (1.0 A) crystal structure of Fusarium solanipisi cutinase: Stereochemical analysis. J. Mol. Biol. 1997, 268, 779–799. [Google Scholar] [CrossRef] [PubMed]
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 1994, 15, 293–308. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, K.E.; Ransac, S.; Dijkstra, B.W.; Colson, C.; van Heuvel, M.; Misset, O. Bacterial lipases. FEMS Microbiol. Rev. 1994, 15, 29–63. [Google Scholar] [CrossRef]
- Mohanan, N.; Sharma, P.K.; Levin, D.B. Characterization of an intracellular poly(3-hydroxyalkanoate) depolymerase from the soil bacterium, Pseudomonas putida LS46. Polym. Degrad. Stab. 2020, 175, 109127. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Steinbüchel, A.; Jendrossek, D. Substrate specificities of bacterial polyhydroxyalkanoate depolymerases and lipases: Bacterial lipases hydrolyze poly(omega-hydroxyalkanoates). Appl. Environ. Microbiol. 1995, 61, 3113–3118. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.-J.; Cho, H. Pseudomonas lipases as catalysts in organic synthesis: Specificity of lipoprotein lipase. J. Chem. Soc. Chem. Commun. 1992, 19, 1411–1413. [Google Scholar] [CrossRef] [Green Version]
- Schirmer, A.; Jendrossek, D.; Schlegel, H.G. Degradation of poly(3-hydroxyoctanoic acid) [P(3HO)] by bacteria: Purification and properties of a [P(3HO)] depolymerase from Pseudomonas fluorescens GK13. Appl. Environ. Microbiol. 1993, 59, 1220–1227. [Google Scholar] [CrossRef] [Green Version]
- Gangoiti, J.; Santos, M.; Llama, M.J.; Serra, J.L. Production of chiral (R)-3-hydroxyoctanoic acid monomers catalyzed by Pseudomonas fluorescens GK13 poly(3-hydroxyoctanoic acid) depolymerase. Appl. Environ. Microbiol. 2020, 76, 3554–3560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, V.; de la Pena, F.; García-Hidalgo, J.; de la Mata, I.; García, J.L.; Prietoa, M.A. Identification and biochemical evidence of a medium-chain length polyhydroxyalkanoate depolymerase in the Bdellovibriobacteriovorus predatory hydrolytic arsenal. Appl. Environ. Microbiol. 2012, 78, 6017–6026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Eugenio, L.I.; Escapa, I.F.; Morales, V.; Dinjaski, N.; Galan, B.; García, J.L.; Prieto, M.A. The turnover of medium-chain-length polyhydroxyalkanoates in Pseudomonas putida KT2442 and the fundamental role of PhaZ depolymerase for the metabolic balance. Environ. Microbiol. 2010, 12, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.K.; Mohanan, N.; Sidhu, R.; Levin, D.B. Colonization and degradation of polyhydroxyalkanoates by lipase producing bacteria. Can. J. Microbiol. 2019, 65, 461–475. [Google Scholar] [CrossRef]
- Schirmer, A.; Jendrossek, D. Molecular characterization of the extracellular poly(3 hydroxyoctanoic acid) [P(3HO)] depolymerase gene of Pseudomonas fluorescens GK13 and of its gene product. J. Bacteriol. 1994, 176, 7065–7073. [Google Scholar] [CrossRef] [Green Version]
- Gangoiti, J.; Santos, M.; Prieto, M.A.; de la Mata, I.; Serra, J.L.; Llama, M.J. Characterization of a novel subgroup of extracellular medium-chain-length polyhydroxyalkanoate depolymerases from Actinobacteria. Appl. Environ. Microbiol. 2012, 78, 7229–7237. [Google Scholar] [CrossRef] [Green Version]
- Arpigny, J.L.; Jaeger, K.E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 1999, 343, 177–183. [Google Scholar] [CrossRef]
- Ollis, D.L.; Cheak, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S.M.; Havel, M.; Remington, S.J.; Silman, I.; Schrag, J.; et al. The α/β hydrolase fold. Protein Eng. 1992, 5, 197–211. [Google Scholar] [CrossRef] [Green Version]
- de Eugenio, L.I.; García, P.; Luengo, J.M.; Sanz, J.; San Roma, J.; García, J.L.; Prieto, M.A. Biochemical evidence that PhaZ gene encodes a specific intracellular medium chain length polyhydroxyalkanoate depolymerase in Pseudomonas putida KT2442: Characterization of a paradigmatic enzyme. J. Biol. Chem. 2007, 282, 4951–4962. [Google Scholar] [CrossRef] [Green Version]
- Chartrain, M.; Katz, L.; Marcin, C.; Thien, M.; Smith, S.; Fisher, E.; Goklen, K.; Salmon, P.; Brix, T.; Price, K.; et al. Purification and characterization of a novel bioconverting lipase from Pseudomonasaeruginosa MB 5001. Enzym. Microb. Technol. 1993, 15, 575–580. [Google Scholar] [CrossRef]
- Fox, P.F.; Stepaniak, L. Isolation and some properties of extracellular heat-stable lipases from Pseudomonas fluorescens strain AFT 36. J. Dairy Res. 1983, 50, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Choo, D.-W.; Kurihara, T.; Suzuki, T.; Soda, K.; Esaki, N. A cold-adapted lipase on an Alaskan psychrotroph, Pseudomonas sp. strain B11-1: Gene cloning and enzyme purification and characterization. Appl. Environ. Microbiol. 1998, 64, 486–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, H.; Gao, S.; Han, S.; Cao, S. Purification and characterization of a Pseudomonas sp. lipase and its properties in non-aqueous media. Biotechnol. Appl. Biochem. 1999, 30, 251–256. [Google Scholar] [PubMed]
- Howard, G.T.; Crother, B.; Vicknair, J. Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas chlororaphis. Int. Biodeter. Biodegrad. 2001, 47, 141–149. [Google Scholar] [CrossRef]
- Adams, D.M.; Brawley, T.G. Heat resistant bacterial lipases and ultra-high temperature sterilization of dairy products. J. Dairy Sci. 1981, 64, 1951–1957. [Google Scholar] [CrossRef]
- Petukhov, M.; Kil, Y.; Kuramitsu, S.; Lanzov, V. Insights into thermal resistance of proteins from the intrinsic stability of their alpha-helices. Proteins 1997, 29, 309–320. [Google Scholar] [CrossRef]
- Gao, X.; Liu, Z.; Cui, W.; Zhou, L.; Tian, Y.; Zhou, Z. Enhanced thermal stability and hydrolytic ability of Bacillus subtilis aminopeptidase by removing the thermal sensitive domain in the non-catalytic region. PLoS ONE 2014, 9, e92357. [Google Scholar] [CrossRef]
- Dharmsthiti, S.; Ammaranond, P. Purification and characterization of lipase from a raw-milk yeast (Trichosporonasteroides). Biotechnol. Appl. Biochem. 1996, 26, 111–116. [Google Scholar]
- Yadav, R.P.; Saxena, R.K.; Gupta, R.; Davidson, S. Purification and characterization of a regiospecific lipase from Aspergillus terreus. Biotechnol. Appl. Biochem. 1998, 28, 243–249. [Google Scholar]
- Lee, S.Y.; Rhee, J.S. Hydrolysis of triglyceride by the whole cell of Pseudomonas putida 3SK in two-phase batch and continuous reactor systems. Biocatal. Bioeng. 1994, 44, 437–443. [Google Scholar] [CrossRef]
- Gilbert, E.J.; Cornish, A.; Jones, C.W. Purification and properties of extracellular lipase from Pseudomonas aeruginosa EF2. J. Gen. Microbiol. 1991, 137, 2223–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.W.; Zeng, R.Y. Molecular cloning and expression of a cold-adapted lipase gene from an Antarctic deep sea psychrotrophic bacterium Pseudomonas sp. 7323. Mar. Biotechnol. 2008, 10, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Brockman, H.L. Dipole potential of lipid membranes. Chem. Phys. Lipids. 1994, 73, 57–79. [Google Scholar] [CrossRef]
- Macrae, A.R.; Hammond, R.C. Present and future applications of lipases. Biotechnol. Genet. Eng. Rev. 1985, 3, 193–218. [Google Scholar] [CrossRef]
- Godtfredsen, S.E. Microbial Lipases. In Microbial Enzymes and Biotechnology; Fogarty, W.M., Kelly, E.T., Eds.; Elsevier: Amsterdam, The Netherlands, 1990; pp. 255–274. [Google Scholar]
- Brocca, S.; Secundo, F.; Ossola, M.; Alberghina, L.; Carrea, G.; Lotti, M. Sequence of the lid affects activity and specificity of Candida rugosa lipase isoenzymes. Protein Sci. 2003, 12, 2312–2319. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Barbe, S.; Lafaquiere, V.; Guieysse, D.; Monsan, P.; Remaud-Siméon, M.; Andre, I. Insights into lid movements of Burkholderiacepacia lipase inferred from molecular dynamics simulations. Proteins 2009, 77, 509–523. [Google Scholar] [CrossRef]
- Yang, Y.; Lowe, M.E. The open lid mediates pancreatic lipase function. J. Lipid Res. 2000, 41, 48–57. [Google Scholar] [CrossRef]
- Holmquist, M.; Clausen, I.G.; Patkar, S.; Svendsen, A.; Hult, K. Probing a functional role of Glu87 and Trp89 in the lid of Humicolalanuginosa lipase through transesterification reactions in organic solvent. J. Protein Chem. 1995, 14, 217–224. [Google Scholar] [CrossRef]
- Kanmani, P.; Kumaresan, K.; Aravind, J.; Karthikeyan, S.; Balan, R. Enzymatic degradation of polyhydroxyalkanoate using lipase from Bacillus subtilis. Int. J. Environ. Sci. Technol. 2016, 13, 1541–1552. [Google Scholar] [CrossRef] [Green Version]
- Mohanan, N.; Wong, C.H.; Budisa, N.; Levin, D.B. Characterization of Polymer Degrading Lipases, LIP1 and LIP2 from Pseudomonas chlororaphis PA23. Front. Bioeng. Biotechnol. 2022, 10, 854298. [Google Scholar] [CrossRef]
- Madison, L.L.; Huisman, G.W. Metabolic engineering of poly(3-hydroxyalkanoates): From DNA to plastic. Microbiol. Mol. Biol. Rev. 1999, 63, 21–53. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Du, P.; Zhong, J.; Bao, Y.; Xu, Z.; Lu, J.; Yang Li, Y. Supervariate ceramics: Biomineralization mechanism. Mater. Today Adv. 2021, 10, 100144. [Google Scholar] [CrossRef]
- Jendrossek, D. Peculiarities of PHA Granules preparation and PHA depolymerase activity determination. Appl. Microbiol. Biotechnol. 2007, 74, 1186–1196. [Google Scholar] [CrossRef] [PubMed]
- Fatma, F.A.; Magdi, A.E.; Soadm, A.E.; Shin-ichi, I. Biodegradation of poly(-caprolactone) (PCL) film and foam plastic by Pseudozyma japonica sp. nov., a novel cutinolyticustilaginomycetous yeast species. Biotech 2014, 4, 507–512. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.; Gangoiti, J.; Keul, H.; Moller, M.; Serra, J.L.; Llama, M.J. Polyester hydrolytic and synthetic activity catalysed by the medium-chain-length poly(3-hydroxyalkanoate) depolymerase from Streptomyces venezuelae SO1. Appl. Microbiol. Biotechnol. 2013, 97, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, R.A.; Salleh, A.B.; Leow, A.T.C.; Yahaya, N.M.; Rahman, M.B.A. Ability of T1 lipase to degrade amorphous P(3HB): Structural and functional study. Mol. Biotechnol. 2017, 59, 284–293.72. [Google Scholar] [CrossRef]
- Kim, D.Y.; Nam, J.S.; Rhee, Y.H. Characterization of an extracellular medium chain-length poly(3-hydroxyalkanoate) depolymerase from Pseudomonas alcaligenes LB19. Biomacromolecules 2002, 3, 291–296. [Google Scholar] [CrossRef]
- Kim, D.Y.; Kim, H.C.; Kim, S.Y.; Rhee, Y.H. Molecular characterization of extracellular medium-chain-length poly(3-hydroxyalkanoate) depolymerase genes from Pseudomonas alcaligenes strains. J. Microbiol. 2005, 43, 285–294. [Google Scholar]
- Rhee, Y.H.; Kim, Y.H.; Shin, K.-S. Characterization of an extracellular poly(3-hydroxyoctanoate) depolymerase from the marine isolate, Pseudomonas luteola M13-4. Enzym. Microb. Technol. 2006, 38, 529–535. [Google Scholar] [CrossRef]
- Ansari, N.F.; Amirul, A.A. Preparation and characterization of polyhydroxyalkanoates macroporous scaffold through enzyme-mediated modifications. Appl. Biochem. Biotechnol. 2013, 170, 690–709. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Jiang, H.; Su, T.; Wang, Z. Purification and characterization of two extracellular polyhydroxyalkanoate depolymerases from Pseudomonas mendocina. Biotechnol. Lett. 2013, 35, 1919–1924. [Google Scholar] [CrossRef] [PubMed]
- Azami, N.A.; Wirjon, I.A.; Kannusamy, S.; The, A.H.; Abdullah, A.A. Enhanced degradation of polyhydroxyalkanoates (PHAs) by newly isolated Burkholderiacepacia DP1 with high depolymerase activity. 3 Biotech 2017, 7, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.K.; Fu, J.; Cicek, N.; Sparling, R.; Levin, D.B. Kinetics of medium-chain length polyhydroxyalkanoate production by a novel isolate of Pseudomonas putida LS46. Can. J. Microbiol. 2011, 58, 982–989. [Google Scholar] [CrossRef]
- Blunt, W.; Dartiailh, C.; Sparling, R.; Gapes, D.; Levin, D.B.; Cicek, N. Carbon flux to growth or polyhydroxyalkanoate synthesis under microaerophilic conditions is affected by fatty acid chain-length in Pseudomonas putida LS46. Appl. Microbiol. Biotechnol. 2018, 102, 6437–6449. [Google Scholar] [CrossRef]
- Ramsay, B.A.; Saracovan, I.; Ramsay, J.L.; Marchessauit, R.H. A Method for the isolation of microorganisms producing extracellular long-side-chain poly (β-3-hydroxyalkanoate) depolymerase. J. Environ. Polym. Degrad. 1994, 2, 1–7. [Google Scholar] [CrossRef]
- Sharma, P.K.; Fu, J.; Spicer, V.; Krokhin, O.V.; Cicek, N.; Sparling, R.; Levin, D.B. Global changes in the proteome of Cupriavidusnecator H16 during poly(3-hydroxybutyrate) synthesis from various biodiesel by-product substrates. AMB Express 2016, 6, 36. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Korman, T.P.; Bowie, J.U. Crystal structure of Proteus Mirabilis lipase, a novel lipase from the proteus/psychrophilic subfamily of Lipase Family I.1. PLoS ONE 2012, 7, e52890. [Google Scholar] [CrossRef] [Green Version]
- Shang, F.; Xu, Y. Crystal structure of a novel esterase CinB from Enterobacter asburiae. Natl. Nat. Sci. Found. China 2019. [Google Scholar] [CrossRef]
- Shang, F.; Lan, J.; Liu, W.; Chen, Y.; Wang, L.; Zhao, J.; Chen, J.; Gao, P.; Ha, N.-C.; Quan, C.; et al. Structural and functional analyses of the lipase CinB from Enterobacter asburiae. Biochem. Biophys. Res. Comm. 2019, 519, 274–279. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with Alpha-fold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. Alpha-fold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformat. 2012, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Rappe, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A., III; Skiff, W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Kouker, G.; Jaeger, K.-E. Specific and sensitive plate assay for bacterial lipases. Appl. Environ. Microbiol. 1987, 53, 211–213. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1996, 72, 248–254. [Google Scholar] [CrossRef]










| Lipases | Accession Number | Location | Gene Size (bp) | Number of Amino Acids (aa) | Molecular Weight (kDa) |
|---|---|---|---|---|---|
| LIP1 | EY04_08410 | Intracellular | 828 | 275 | 29.7 |
| LIP2 | EY04_09635 | Intracellular | 834 | 277 | 30.2 |
| LIP3 | EY04_02420 | Intracellular | 891 | 296 | 32.5 |
| LIP4 | EY04_21540 | Extracellular | 942 | 315 | 34.0 |
| LIP5 | EY04_17885 | Extracellular | 1905 | 634 | 69.8 |
| LIP6 | EY04_32435 | Extracellular | 1911 | 636 | 70 |
| PhaZ | EY04_01535 | Intracellular | 858 | 285 | 31.7 |
| Modulators/Reagents | Final Concentration | Relative Activity (%) * | ||
|---|---|---|---|---|
| LIP3 | LIP4 | PhaZ | ||
| EDTA | 10 mmol L−1 | 94 ± 1.0 | 98 ± 1.8 | 97 ± 0.9 |
| CaCl2 | 1 mmol L−1 | 93 ± 2.5 | 91 ± 3.3 | 94 ± 1.6 |
| MgCl2 | 1 mmol L−1 | 96 ± 2.1 | 94 ± 2.6 | 74 ± 2.7 |
| NaCl | 50 mmol L−1 | 90 ± 3.8 | 89 ± 3.1 | 78 ± 3.6 |
| PMSF | 1 mmol L−1 | 71 ± 4.5 | 67 ± 4.5 | 59 ± 3.1 |
| Tween80 | 0.1% | 47 ± 3.8 | 36 ± 3.5 | 53 ± 2.3 |
| SDS | 5% | 30 ± 0.6 | 34 ± 1.5 | 28 ± 1.1 |
| Enzyme activity (without modulator) | 100 | 100 | 100 | |
| Substrate | Relative Activity (%) * | ||
|---|---|---|---|
| LIP3 | LIP4 | PhaZ | |
| PHA polymers | |||
| PHBV | 70.82 ± 3.6 | 48.78 ± 2.9 | 43.64 ± 5.1 |
| PHHx | 100 | 100 | 76.70 ± 3.6 |
| PHO | 57.40 ± 1.9 | 86.25 ± 5.2 | 100 |
| PHN | 54.52 ± 4.6 | 74.92 ± 1.6 | 91.86 ± 4.2 |
| PHD | 52.07 ± 5.2 | 61.44 ± 4.7 | 88.23 ± 2.9 |
| Other biodegradable/petrochemical-based polymers | |||
| Polylactic acid (PLA) | 53.5 ± 2.7 | 33.16 ± 2.1 | 40.42 ± 3.7 |
| Poly(ε-caprolactone) (PCL) | 25.43 ± 3.9 | 47.65 ± 2.6 | 38.90 ± 4.1 |
| Poly(ethylene succinate) (PES) | 13.21 ± 4.1 | 11.32 ± 4.1 | 22.76 ± 4.1 |
| Polymer Substrate | Mw before (Da) | Mw after (Da) | Mw (Da) | % Change |
|---|---|---|---|---|
| PHBV | 34,542 | 28,043 | 6499 | 18.8 |
| PHHHx | 27,177 | 20,820 | 6357 | 23.4 |
| PHO | 21,477 | 17,157 | 4320 | 20.1 |
| PHN | 58,889 | 34,907 | 23,982 | 49.7 |
| PHD | 27,490 | 20,348 | 7142 | 25.9 |
| PLA | 191,677 | 148,292 | 43,285 | 22.5 |
| PCL | 186,692 | 134,503 | 52,189 | 27.9 |
| PES | 3619 | 3381 | 238 | 6.6 |
| Polymer Substrate | Mw before (Da) | Mw after (Da) | Mw (Da) | % Change |
|---|---|---|---|---|
| PHBV | 34,542 | 27,687 | 6855 | 19.8 |
| PHHHx | 27,177 | 21,234 | 5943 | 21.9 |
| PHO | 21,477 | 18,106 | 3371 | 15.7 |
| PHN | 58,889 | 49,711 | 9178 | 15.6 |
| PHD | 27,490 | 21,590 | 5900 | 21.5 |
| PLA | 191,677 | 174,894 | 16,783 | 8.7 |
| PCL | 186,692 | 94,663 | 92,092 | 49.3 |
| PES | 3619 | 3414 | 205 | 5.7 |
| Polymer Substrate | Mw before (Da) | Mw after (Da) | Mw (Da) | % Change |
|---|---|---|---|---|
| PHBV | 34,542 | 29,479 | 5063 | 14.7 |
| PHHHx | 27,177 | 23,052 | 4125 | 15.2 |
| PHO | 21,477 | 20,146 | 1331 | 6.2 |
| PHN | 58,889 | 50,418 | 8471 | 14.4 |
| PHD | 27,490 | 24,302 | 3188 | 11.6 |
| PLA | 191,677 | 163,042 | 28,635 | 14.9 |
| PCL | 186,692 | 135,128 | 51,564 | 27.6 |
| PES | 3619 | 3283 | 336 | 9.3 |
| Bacterial Source | Lipases/ Esterases | Polyesters Hydrolyzed/Degradation Ability of the Enzymes | Polyesters NotHydrolyzed | Method Used | Activity | Ref. |
|---|---|---|---|---|---|---|
| Bacillus subtilis, Pseudomonas aeruginosa, Pseudomonas alcaligenes, Burkholderiacepacia (former Pseudomonas cepacia) | Extracellular lipases | pNP-palmitate Poly(6-hydroxyhexanoate) Poly(4-hydroxybutyrate) Polycaprolactone | Poly(3-hydroxybutyrate) Poly(3-hydroxyalkanoates) Polylactic acid | Turbidimetric assay; Rhodamine agar plate assay. | B. subtilis lipase: 0.2 × 103 U/mg (pNPP) 6000 U mg−1 (PCL) P. aeruginosa lipase: 52 × 103 U mg−1 (pNPP) 1.8 × 106 U mg−1 (PCL) P. alcaligenes lipase: 8 × 103 U mg−1 (pNPP) 140,000 U mg−1 (PCL) B. cepacia lipase: 0.5 × 103 U mg−1 (pNPP) 40,000 U mg−1 (PCL) | [18] |
| P. fluorescens GK13 | Extracellular esterase | vvpNPP | Polyhydroxyalkanoates Polycaprolactone Polylactic acid | Turbidimetric assay; Rhodamine agar plate assay. | 0.4 × 103 U mg−1 (pNPP) | [18] |
| Bacillus subtilis | Extracellular lipase | pNPP Polyhydroxyalkanoates | - | Molecular weight decrease by GPC; FTIR; NMR; DSC. | 21.3% molecular weight decrease 28.3% weight loss 273.65 U mg−1 (pNPP) | [51] |
| Geobacilluszalihae | Extracellular lipase | Poly(3-hydroxybutyrate) | - | Poly(3-hydroxybutyrate) agar plate; | Clear zone around the colony | [58] |
| Pseudomonas chlororaphis PA23-63-1 | Extracellular lipases and esterases mix | pNP-alkanoate Poly (3-hydroxybutyrate-co-3-hydroxyvalerate) Poly(3-hydroxyhexanoate) Poly(3-hydroxyoctanoate) Poly(3-hydroxynonanoate) Poly(3-hydroxydecanoate) | Polycaprolactone Polyethylene sulfonate | PHA agar plate assay; Turbidimetric assay; Weight loss of PHA films. | 4.5% weight loss 997.7 U mg−1 (pNPO) 722 U mg−1 (PHO) Clear zone of PHA hydrolysis | [24] |
| Pseudomonas chlororaphis PA23 | Intracellular lipases, LIP1, and LIP2 | Poly (3-hydroxybutyrate-co-3-hydroxyvalerate Poly(3-hydroxyhexanoate) Poly(3-hydroxyoctanoate) Poly(3-hydroxynonanoate) Poly(3-hydroxydecanoate) Polylactic acid Polycaprolactone Polyethylene sulfonate | - | Nile blue agar plate assay; Turbidimetric assay; Molecular weight decrease by GPC. | 18–40% molecular weight decrease Clear zone of PHA hydrolysis 769.23 U mg− 1 in LIP1 (pNPO) 714.28 U mg−1 in LIP1 (pNPO) 360.12 U mg−1 in LIP1 (PHO) 301.72 U mg−1 in LIP2 (PHO) | [52] |
| Pseudomonas alcaligenes LB19 | Extracellular PHA depolymerase | pNP-alkanoate Poly(3-hydroxydecanoate) | Poly(3-hydroxybutyrate) Polycaprolactone Poly(L-lactide) | Monomer composition of hydrolysis products by GC chromatography. | 2000 U mg−1 (PHO) | [59,60] |
| Pseudomonas luteola M13-4 | Extracellular PHA depolymerase | pNP-alkanoate Poly(3-hydroxyalkanoate) Poly (3-hydroxybutyrate-co-3-hydroxyvalerate Poly(3-hydroxyoctanoate) Poly(3-hydroxyheptanoate) Poly(3-hydroxydodecanoate) | Poly(3-hydroxybutyrate) Polycaprolactone | Turbidimetric assay. | 2.21 U mg−1 (Poly(3-hydroxydodecanoate) | [61] |
| Bdellovibriobacteriovorus HD100 | Extracellular PHA depolymerase | Poly-(hydroxyoctanoate-cohydroxyhexanoate) | Poly(3-hydroxybutyrate) | HPLC-MS analysis of degradation products. | 55 ± 2 U mg−1 (PHA) | [22] |
| Acidovorax sp. DP5 | Extracellular PHA depolymerase | Poly-(3-hydroxybutyrate-co-4-hydroxybutyrate) [P(3HB-co-70%4HB)] | - | Weight loss. | 8% weight loss 0.0075 U ml−1 (PHA) | [62] |
| Pseudomonas mendocina DS04-T | Extracellular Polyhydroxyalkanoate depolymerases, PHAase I, and PHAase II | Polyhydroxybutyrate Poly (3-hydroxybutyrate-co-3-hydroxyvalerate Poly-(3-hydroxybutyrate-co-4-hydroxybutyrate) [P(3HB-co-4HB)] | Poly(3-hydroxybutyrate) Polylactic acid by PHAse I | Weight loss. | 1899 U mg−1 (PHA) (PHAase I) 893 U mg−1 (PHA) (PHAase II) | [63] |
| Pseudomonas putida | Intracellular PHA depolymerase, PhaZ | Poly (3-hydroxybutyrate-co-3-hydroxyvalerate Poly(3-hydroxyhexanoate) Poly(3-hydroxyoctanoate) Poly(3-hydroxynonanoate) Poly(3-hydroxydecanoate) Polylactic acid Polycaprolactone Polyethylene sulfonate | - | Nile blue agar plate assay; Turbidimetric assay; Molecular weight decrease by GPC. | Molecular weight decrease Clear zone of PHA hydrolysis 172.8 U mg−1 (PHO) | [17] |
| Burkoldariacepacia DP1 | Extracellular PHA depolymerase | Poly-(3-hydroxybutyrate-co-4-hydroxybutyrate) [P(3HB-co-21%4HB)] | - | Weight loss. | - | [64] |
| Pseudomonas chlororaphis PA23 | Intracellular lipase, LIP3 Extracellular lipase, LIP4 | Poly (3-hydroxybutyrate-co-3-hydroxyvalerate Poly(3-hydroxyhexanoate) Poly(3-hydroxyoctanoate) Poly(3-hydroxynonanoate) Poly(3-hydroxydecanoate) Polylactic acid Polycaprolactone Polyethylene sulfonate | - | Nile blue agar plate assay; Turbidimetric assay; Molecular weight decrease by GPC. | 49.3–49.7% molecular weight decrease Clear zone of PHA hydrolysis 625.0 U mg−1 in LIP3 (pNPO) 555.55 U mg−1 in LIP4 (pNPO) 122.13 U mg−1 in LIP3 (PHO) 102.6 U mg−1 in LIP4 (PHO) | This study |
| Pseudomonas chlororaphis PA23 | Intracellular PHA depolymerase, PhaZ | Poly (3-hydroxybutyrate-co-3-hydroxyvalerate Poly(3-hydroxyhexanoate) Poly(3-hydroxyoctanoate) Poly(3-hydroxynonanoate) Poly(3-hydroxydecanoate) Polylactic acid Polycaprolactone Polyethylene sulfonate | - | Nile blue agar plate assay; Turbidimetric assay; Molecular weight decrease by GPC. | 27.6% molecular weight decrease Clear zone of PHA hydrolysis 909.09 U mg−1 (pNPO) 650.13 U mg−1 (PHO) | This study |
| PHA * | Substrate | * Monomer Composition of PHA (mol%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3HB | 3HV | 3HHx | 3HHp | 3HO | 3HN | 3HD | 3HDD | 3HTD | ||
| PHBV | Glucose/ Valerate | 76.9 | 23.1 | ND | ND | ND | ND | ND | ND | ND |
| PHHx | Hexanoic acid | ND | ND | 82.4 | ND | 16.0 | ND | 1.6 | ND | ND |
| PHO | Octanoic acid | ND | ND | 6.5 | ND | 92.0 | ND | 1.5 | ND | ND |
| PHN | Nonanoic acid | ND | ND | ND | 18.7 | ND | 81.3 | ND | ND | ND |
| PHD | Decanoic acid | ND | ND | 5.2 | ND | 57.4 | ND | 37.0 | 0.4 | ND |
| Gene | Primers Used |
|---|---|
| lip3 | FP: 5′-ATAGCTAGCTCCCAAGAGCTTGCCACGCGT-3′ |
| RP: 5′-ATAAAGCTTTACCCCCGCCGCTTTCAATCG-3′ | |
| lip4 | FP: 5′-ATAGCTAGCGGGGTCGAACACAACACCCAG-3′ |
| RP: 5′-CCAAGCTTCTTCAGGTGTTGCTTGAGCTCT-3′ | |
| phaZ | FP: 5′-ATAGCTAGCCCTCAACCGTTCATCTTTCGC-3′ |
| RP: 5′-CCAAGCTTCGTACCGCTGAACGGTGTCGGA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohanan, N.; Wong, M.C.-H.; Budisa, N.; Levin, D.B. Polymer-Degrading Enzymes of Pseudomonas chloroaphis PA23 Display Broad Substrate Preferences. Int. J. Mol. Sci. 2023, 24, 4501. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms24054501
Mohanan N, Wong MC-H, Budisa N, Levin DB. Polymer-Degrading Enzymes of Pseudomonas chloroaphis PA23 Display Broad Substrate Preferences. International Journal of Molecular Sciences. 2023; 24(5):4501. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms24054501
Chicago/Turabian StyleMohanan, Nisha, Michael C.-H. Wong, Nediljko Budisa, and David B. Levin. 2023. "Polymer-Degrading Enzymes of Pseudomonas chloroaphis PA23 Display Broad Substrate Preferences" International Journal of Molecular Sciences 24, no. 5: 4501. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms24054501