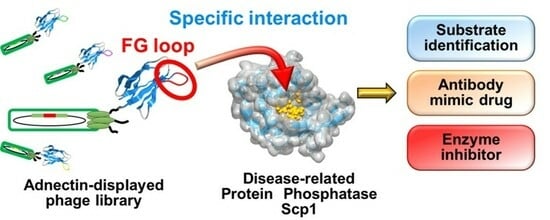
Identification of Inhibitors of the Disease-Associated Protein Phosphatase Scp1 Using Antibody Mimetic Molecules
Abstract
:1. Introduction
2. Results
2.1. Construction of an Adnectin-Derived Phage Display Library with a Randomized FG Loop
2.2. Screening of Scp1-Specific Adnectin from the Constructed Phage Display Library
2.3. In Vitro Functional Analysis of Recombinant Scp1-Specific Adnectin
2.4. Biological Functions of FG-1Adn in Cells
3. Discussion
4. Materials and Methods
4.1. Preparation of an Adnectin-Derived Phage Library
4.2. Screening of Adnectins against Scp1 Using FG loop Randomized Adnectin-Derived Phage Display Library
4.3. Expression and Purification of Recombinant Adnectins
4.4. Binding Analysis of Adnectin-Derived Phages
4.5. Binding Analysis of Peptide-Conjugated Bacterial Alkaline Phosphatase
4.6. Binding Analysis of Recombinant Adnectins
4.7. pNPP Phosphatase Assay
4.8. Malachite Green Assay
4.9. Biolayer Interferometry BLItz System Assay
4.10. Subcellular Localization Analysis of Transfected Adnectin
4.11. Western Blotting
4.12. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef]
- Shi, Y. Serine/threonine phosphatases: Mechanism through structure. Cell 2009, 139, 468–484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kim, Y.; Genoud, N.; Gao, J.; Kelly, J.W.; Pfaff, S.L.; Gill, G.N.; Dixon, J.E.; Noel, J.P. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol. Cell 2006, 24, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Yeo, M.; Lin, P.S.; Dahmus, M.E.; Gill, G.N. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J. Biol. Chem. 2003, 278, 26078–26085. [Google Scholar] [CrossRef] [PubMed]
- Harikrishna, R.; Kim, H.; Noh, K.; Kim, Y.J. The diverse roles of RNA polymerase II C-terminal domain phosphatase SCP1. BMB Rep. 2014, 47, 192–196. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, Y.; Zhang, Q.; Liu, J.J.; Li, T.J.; Yang, J.R.; Zeng, C.; Zhuang, S.M. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012, 40, 4615–4625. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, K.H.; Willis, D.; Long, J.; Liu, F.; Lin, X.; Feng, X.H. Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-beta signaling. J. Biol. Chem. 2006, 281, 38365–38375. [Google Scholar] [CrossRef] [PubMed]
- Burkholder, N.T.; Mayfield, J.E.; Yu, X.; Irani, S.; Arce, D.K.; Jiang, F.; Matthews, W.L.; Xue, Y.; Zhang, Y.J. Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST). J. Biol. Chem. 2018, 293, 16851–16861. [Google Scholar] [CrossRef] [PubMed]
- Gervasi, N.M.; Dimtchev, A.; Clark, D.M.; Dingle, M.; Pisarchik, A.V.; Nesti, L.J. C-terminal domain small phosphatase 1 (CTDSP1) regulates growth factor expression and axonal regeneration in peripheral nerve tissue. Sci. Rep. 2021, 11, 14462. [Google Scholar] [CrossRef]
- Liao, P.; Wang, W.; Li, Y.; Wang, R.; Jin, J.; Pang, W.; Chen, Y.; Shen, M.; Wang, X.; Jiang, D.; et al. Palmitoylated SCP1 is targeted to the plasma membrane and negatively regulates angiogenesis. Elife 2017, 6, e22058. [Google Scholar] [CrossRef]
- Yeo, M.; Lee, S.K.; Lee, B.; Ruiz, E.C.; Pfaff, S.L.; Gill, G.N. Small CTD phosphatases function in silencing neuronal gene expression. Science 2005, 307, 596–600. [Google Scholar] [CrossRef]
- Ashton, N.J.; Hye, A.; Leckey, C.A.; Jones, A.R.; Gardner, A.; Elliott, C.; Wetherell, J.L.; Lenze, E.J.; Killick, R.; Marchant, N.L. Plasma REST: A novel candidate biomarker of Alzheimer’s disease is modified by psychological intervention in an at risk population. Transl. Psychiatry 2017, 7, e1148. [Google Scholar] [CrossRef]
- Kawamura, M.; Sato, S.; Matsumoto, G.; Fukuda, T.; Shiba-Fukushima, K.; Noda, S.; Takanashi, M.; Mori, N.; Hattori, N. Loss of nuclear REST/NRSF in aged-dopaminergic neurons in Parkinson’s disease patients. Neurosci. Lett. 2019, 699, 59–63. [Google Scholar] [CrossRef] [PubMed]
- de Souza, J.M.; Abd-Elrahman, K.S.; Ribeiro, F.M.; Ferguson, S.S.G. mGluR5 regulates REST/NRSF signaling through N-cadherin/β-catenin complex in Huntington’s disease. Mol. Brain 2020, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Spencer, E.M.; Chandler, K.E.; Haddley, K.; Howard, M.R.; Hughes, D.; Belyaev, N.D.; Coulson, J.M.; Stewart, J.P.; Buckley, N.J.; Kipar, A.; et al. Regulation and role of REST and REST4 variants in modulation of gene expression in in vivo and in vitro in epilepsy models. Neurobiol. Dis. 2006, 24, 41–52. [Google Scholar] [CrossRef]
- Wu, Y.; Evers, B.M.; Zhou, B.P. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J. Biol. Chem. 2009, 284, 640–648. [Google Scholar] [CrossRef]
- Qian, W.; Li, Q.; Wu, X.; Li, W.; Li, Q.; Zhang, J.; Li, M.; Zhang, D.; Zhao, H.; Zou, X.; et al. Deubiquitinase USP29 promotes gastric cancer cell migration by cooperating with phosphatase SCP1 to stabilize Snail protein. Oncogene 2020, 39, 6802–6815. [Google Scholar] [CrossRef]
- Garcia-Manteiga, J.M.; D’Alessandro, R.; Meldolesi, J. News about the Role of the Transcription Factor REST in Neurons: From Physiology to Pathology. Int. J. Mol. Sci. 2019, 21, 235. [Google Scholar] [CrossRef] [PubMed]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef]
- Liu, T.; Wang, W.; Li, X.; Chen, Y.; Mu, F.; Wen, A.; Liu, M.; Ding, Y. Advances of phytotherapy in ischemic stroke targeting PI3K/Akt signaling. Phytother. Res. 2023, 37, 5509–5528. [Google Scholar] [CrossRef]
- Sharma, A.; Mehan, S. Targeting PI3K-AKT/mTOR signaling in the prevention of autism. Neurochem. Int. 2021, 147, 105067. [Google Scholar] [CrossRef] [PubMed]
- Kanlaya, R.; Kapincharanon, C.; Fong-Ngern, K.; Thongboonkerd, V. Induction of mesenchymal-epithelial transition (MET) by epigallocatechin-3-gallate to reverse epithelial-mesenchymal transition (EMT) in SNAI1-overexpressed renal cells: A potential anti-fibrotic strategy. J. Nutr. Biochem. 2022, 107, 109066. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cho, E.J.; Burstein, G.; Siegel, D.; Zhang, Y. Selective inactivation of a human neuronal silencing phosphatase by a small molecule inhibitor. ACS Chem. Biol. 2011, 6, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Medellin, B.; Yang, W.; Konduri, S.; Dong, J.; Irani, S.; Wu, H.; Matthews, W.L.; Zhang, Z.Y.; Siegel, D.; Zhang, Y. Targeted Covalent Inhibition of Small CTD Phosphatase 1 to Promote the Degradation of the REST Transcription Factor in Human Cells. J. Med. Chem. 2022, 65, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, K.; Yoneda, T.; Kaneko, A.; Yagi, S.; Furukawa, K.; Chuman, Y. Development of a Substrate Identification Method for Human Scp1 Phosphatase Using Phosphorylation Mimic Phage Display. Protein Pept. Lett. 2018, 25, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Yamazaki, K.; Imai, S.; Banno, A.; Kaneko, A.; Furukawa, K.; Chuman, Y. Identification of a Specific Inhibitor of Human Scp1 Phosphatase Using the Phosphorylation Mimic Phage Display Method. Catalysts 2019, 9, 842. [Google Scholar] [CrossRef]
- Mizunuma, M.; Kaneko, A.; Imai, S.; Furukawa, K.; Chuman, Y. Methods for Identification of Substrates/Inhibitors of FCP/SCP Type Protein Ser/Thr Phosphatases. Processes 2020, 8, 1598. [Google Scholar] [CrossRef]
- Williams, M.P.; Pounder, R.E. Review article: The pharmacology of rabeprazole. Aliment. Pharmacol. Ther. 1999, 13, 3–10. [Google Scholar] [CrossRef]
- Saeed, A.F.; Wang, R.; Ling, S.; Wang, S. Antibody Engineering for Pursuing a Healthier Future. Front. Microbiol. 2017, 8, 495. [Google Scholar] [CrossRef]
- Simeon, R.; Chen, Z. In vitro-engineered non-antibody protein therapeutics. Protein Cell. 2018, 9, 3–14. [Google Scholar] [CrossRef]
- Sha, F.; Salzman, G.; Gupta, A.; Koide, S. Monobodies and other synthetic binding proteins for expanding protein science. Protein Sci. 2017, 26, 910–924. [Google Scholar] [CrossRef]
- Liu, M.; Li, L.; Jin, D.; Liu, Y. Nanobody-A versatile tool for cancer diagnosis and therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021, 13, e1697. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/33470555 (accessed on 25 February 2024). [CrossRef]
- Plückthun, A. Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 489–511. [Google Scholar] [CrossRef]
- Rothe, C.; Skerra, A. Anticalin® Proteins as Therapeutic Agents in Human Diseases. BioDrugs 2018, 32, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Bailey, C.W.; Huang, X.; Koide, S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998, 284, 1141–1151. [Google Scholar] [CrossRef]
- Park, S.H.; Park, S.; Kim, D.Y.; Pyo, A.; Kimura, R.H.; Sathirachinda, A.; Choy, H.E.; Min, J.J.; Gambhir, S.S.; Hong, Y. Isolation and Characterization of a Monobody with a Fibronectin Domain III Scaffold That Specifically Binds EphA2. PLoS ONE 2015, 10, e0132976. [Google Scholar] [CrossRef] [PubMed]
- Weidle, U.H.; Auer, J.; Brinkmann, U.; Georges, G.; Tiefenthaler, G. The emerging role of new protein scaffold-based agents for treatment of cancer. Cancer Genom. Proteom. 2013, 10, 155–168. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/23893924 (accessed on 23 February 2024).
- Koide, A.; Wojcik, J.; Gilbreth, R.N.; Hoey, R.J.; Koide, S. Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. J. Mol. Biol. 2012, 415, 393–405. [Google Scholar] [CrossRef]
- Batori, V.; Koide, A.; Koide, S. Exploring the potential of the monobody scaffold: Effects of loop elongation on the stability of a fibronectin type III domain. Protein Eng. 2002, 15, 1015–1020. [Google Scholar] [CrossRef] [PubMed]
- Cheung, L.S.; Shea, D.J.; Nicholes, N.; Date, A.; Ostermeier, M.; Konstantopoulos, K. Characterization of monobody scaffold interactions with ligand via force spectroscopy and steered molecular dynamics. Sci. Rep. 2015, 5, 8247. [Google Scholar] [CrossRef]
- Richards, J.; Miller, M.; Abend, J.; Koide, A.; Koide, S.; Dewhurst, S. Engineered fibronectin type III domain with a RGDWXE sequence binds with enhanced affinity and specificity to human alphavbeta3 integrin. J. Mol. Biol. 2003, 326, 1475–1488. [Google Scholar] [CrossRef] [PubMed]
- Karatan, E.; Merguerian, M.; Han, Z.; Scholle, M.D.; Koide, S.; Kay, B.K. Molecular recognition properties of FN3 monobodies that bind the Src SH3 domain. Chem. Biol. 2004, 11, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Ikeura, M.; Tashiro, H.; Yamagata, Y.; Saito, H.; Kobayashi, T.; Mizunuma, M.; Yamazaki, K.; Baba, K.; Furukawa, K.; Chuman, Y. Development of Antibody-like Proteins Targeting the Oncogenic Ser/Thr Protein Phosphatase PPM1D. Processes 2022, 10, 1501. [Google Scholar] [CrossRef]
- Guntas, G.; Lewis, S.M.; Mulvaney, K.M.; Cloer, E.W.; Tripathy, A.; Lane, T.R.; Major, M.B.; Kuhlman, B. Engineering a genetically encoded competitive inhibitor of the KEAP1-NRF2 interaction via structure-based design and phage display. Protein Eng. Des. Sel. 2016, 29, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wallon, L.; Khan, I.; Teng, K.W.; Koide, A.; Zuberi, M.; Li, J.; Ketavarapu, G.; Traaseth, N.J.; O’Bryan, J.P.; Koide, S. Inhibition of RAS-driven signaling and tumorigenesis with a pan-RAS monobody targeting the Switch I/II pocket. Proc. Natl. Acad. Sci. USA 2022, 119, e2204481119. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Iwatani, Y.; Matsuoka, K.; Fujino, T.; Umemoto, S.; Yokomaku, Y.; Ishizaki, K.; Kito, S.; Sezaki, T.; Hayashi, G.; et al. Antibody-like proteins that capture and neutralize SARS-CoV-2. Sci. Adv. 2020, 6, eabd3916. [Google Scholar] [CrossRef] [PubMed]
- Rallabandi, H.R.; Ganesan, P.; Kim, Y.J. Targeting the C-Terminal Domain Small Phosphatase 1. Life 2020, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Kamenski, T.; Heilmeier, S.; Meinhart, A.; Cramer, P. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol. Cell 2004, 15, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, J.; Lamontanara, A.J.; Grabe, G.; Koide, A.; Akin, L.; Gerig, B.; Hantschel, O.; Koide, S. Allosteric Inhibition of Bcr-Abl Kinase by High Affinity Monobody Inhibitors Directed to the Src Homology 2 (SH2)-Kinase Interface. J. Biol. Chem. 2016, 291, 8836–8847. [Google Scholar] [CrossRef]
- Wang, W.; Liao, P.; Shen, M.; Chen, T.; Chen, Y.; Li, Y.; Lin, X.; Ge, X.; Wang, P. SCP1 regulates c-Myc stability and functions through dephosphorylating c-Myc Ser62. Oncogene 2016, 35, 491–500. [Google Scholar] [CrossRef]
- Song, Z.; Zhao, D.; Zhao, H.; Yang, L. NRSF: An angel or a devil in neurogenesis and neurological diseases. J. Mol. Neurosci. 2015, 56, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Palm, K.; Metsis, M.; Timmusk, T. Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat. Brain Res. Mol. Brain Res. 1999, 72, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Zuccato, C.; Tartari, M.; Crotti, A.; Goffredo, D.; Valenza, M.; Conti, L.; Cataudella, T.; Leavitt, B.R.; Hayden, M.R.; Timmusk, T.; et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003, 35, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Ngambenjawong, C.; Pineda, J.M.; Pun, S.H. Engineering an Affinity-Enhanced Peptide through Optimization of Cyclization Chemistry. Bioconjug. Chem. 2016, 27, 2854–2862. [Google Scholar] [CrossRef]
- Mihara, E.; Watanabe, S.; Bashiruddin, N.K.; Nakamura, N.; Matoba, K.; Sano, Y.; Maini, R.; Yin, Y.; Sakai, K.; Arimori, T.; et al. Lasso-grafting of macrocyclic peptide pharmacophores yields multi-functional proteins. Nat. Commun. 2021, 12, 1543. [Google Scholar] [CrossRef] [PubMed]
- Mamluk, R.; Carvajal, I.M.; Morse, B.A.; Wong, H.; Abramowitz, J.; Aslanian, S.; Lim, A.C.; Gokemeijer, J.; Storek, M.J.; Lee, J.; et al. Anti-tumor effect of CT-322 as an adnectin inhibitor of vascular endothelial growth factor receptor-2. MAbs 2010, 2, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Schiff, D.; Kesari, S.; de Groot, J.; Mikkelsen, T.; Drappatz, J.; Coyle, T.; Fichtel, L.; Silver, B.; Walters, I.; Reardon, D. Phase 2 study of CT-322, a targeted biologic inhibitor of VEGFR-2 based on a domain of human fibronectin, in recurrent glioblastoma. Investig. New Drugs 2015, 33, 247–253. [Google Scholar] [CrossRef]
- Mitchell, T.; Chao, G.; Sitkoff, D.; Lo, F.; Monshizadegan, H.; Meyers, D.; Low, S.; Russo, K.; DiBella, R.; Denhez, F.; et al. Pharmacologic profile of the Adnectin BMS-962476, a small protein biologic alternative to PCSK9 antibodies for low-density lipoprotein lowering. J. Pharmacol. Exp. Ther. 2014, 350, 412–424. [Google Scholar] [CrossRef]
- Yu, X.; Yang, Y.P.; Dikici, E.; Deo, S.K.; Daunert, S. Beyond Antibodies as Binding Partners: The Role of Antibody Mimetics in Bioanalysis. Annu. Rev. Anal. Chem. 2017, 10, 293–320. [Google Scholar] [CrossRef]
- Futaki, S.; Nakase, I. Cell-Surface Interactions on Arginine-Rich Cell-Penetrating Peptides Allow for Multiplex Modes of Internalization. Acc. Chem. Res. 2017, 50, 2449–2456. [Google Scholar] [CrossRef]
- Habault, J.; Poyet, J.L. Recent Advances in Cell Penetrating Peptide-Based Anticancer Therapies. Molecules 2019, 24, 927. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Lu, L.T.; Chen, H.Y.; Duan, X.; Lin, X.; Feng, X.H.; Tang, M.J.; Chen, R.H. SCP phosphatases suppress renal cell carcinoma by stabilizing PML and inhibiting mTOR/HIF signaling. Cancer Res. 2014, 746, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, G.S.; Puzanov, G.A.; Dashinimaev, E.B.; Vishnyakova, K.S.; Kondratieva, T.T.; Chegodaev, Y.S.; Postnov, A.Y.; Senchenko, V.N.; Yegorov, Y.E. Tumor Suppressor Properties of Small C-Terminal Domain Phosphatases in Clear Cell Renal Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 12986. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.L.; Li, S.M.; Hou, J.Y.; Hong, Y.H.; Chen, X.X.; Zhou, C.Q.; Wu, H.; Zheng, G.H.; Zeng, C.T.; Wu, H.D.; et al. Elabela, a Novel Peptide, Exerts Neuroprotective Effects Against Ischemic Stroke Through the APJ/miR-124-3p/CTDSP1/AKT Pathway. Cell. Mol. Neurobiol. 2023, 43, 2989–3003. [Google Scholar] [CrossRef] [PubMed]








| Name | Sequence | Frequency | |
|---|---|---|---|
| 3rd Round | 4th Round | ||
| FG-1 | FVFGPDA | 1 | 12 |
| FG-2 | LLEPEDW | 1 | 4 |
| FG-3 | RPESWGI | 1 | |
| FG-4 | TFQCSIL | 1 | |
| FG-5 | LFEPADR | 1 | |
| Total | 3 | 18 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobayashi, T.; Yamazaki, K.; Shinada, J.; Mizunuma, M.; Furukawa, K.; Chuman, Y. Identification of Inhibitors of the Disease-Associated Protein Phosphatase Scp1 Using Antibody Mimetic Molecules. Int. J. Mol. Sci. 2024, 25, 3737. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms25073737
Kobayashi T, Yamazaki K, Shinada J, Mizunuma M, Furukawa K, Chuman Y. Identification of Inhibitors of the Disease-Associated Protein Phosphatase Scp1 Using Antibody Mimetic Molecules. International Journal of Molecular Sciences. 2024; 25(7):3737. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms25073737
Chicago/Turabian StyleKobayashi, Tamaki, Kazuki Yamazaki, Junki Shinada, Masataka Mizunuma, Kazuhiro Furukawa, and Yoshiro Chuman. 2024. "Identification of Inhibitors of the Disease-Associated Protein Phosphatase Scp1 Using Antibody Mimetic Molecules" International Journal of Molecular Sciences 25, no. 7: 3737. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms25073737