Restoration of the Genus Paraunisaccoides Martin, 1973 (Digenea: Haploporidae) and Description of P. elegans n. sp. and Unisaccus halongi n. sp. from Mugilid Fish in Vietnam
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Trematodes
2.2. DNA Extraction, Amplification and Sequencing
2.3. Alignments and Phylogenetic Analysis
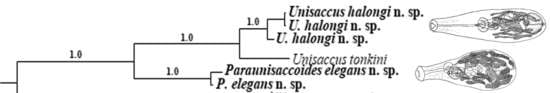
3. Results
3.1. Diagnosis of the Genus Paraunisaccoides
3.2. Paraunisaccoides elegans n. sp.
3.2.1. Taxonomic Summary
3.2.2. Morphology
3.2.3. Molecular Data
3.3. Unisaccus halongi n. sp.
3.3.1. Taxonomic Summary
3.3.2. Morphology
3.3.3. Molecular Data
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Overstreet, R.M.; Curran, S.S. Family Haploporidae Nicoll, 1914. In Keys to the Trematoda; Jones, A., Bray, R.A., Gibson, D.I., Eds.; CABI: Cambridge, MA, USA, 2005; Volume 2, pp. 129–165. [Google Scholar]
- Pulis, E.E.; Overstreet, R.M. Review of haploporid (Trematoda) genera with ornate muscularization in the region of the oral sucker, including four new species and a new genus. Syst. Parasitol. 2013, 84, 167–191. [Google Scholar] [CrossRef]
- Andres, M.J.; Pulis, E.E.; Cribb, T.H.; Overstreet, R.M. Erection of the haploporid genus Litosaccus n.g. and its phylogenetic relationship within the Haploporidae Nicoll, 1914. Syst. Parasitol. 2014, 89, 185–194. [Google Scholar] [CrossRef]
- Besprozvannykh, V.V.; Atopkin, D.M.; Ermolenko, A.V.; Nikitenko, A.Y. Restoration of the genus Parasaccocoelium Zhukov, 1971 (Digenea: Haploporidae) and a description of two new species from mugilid fish in the Far East of Russia. J. Helminthol. 2015, 89, 565–576. [Google Scholar] [CrossRef] [Green Version]
- Besprozvannykh, V.V.; Atopkin, D.M.; Ermolenko, A.V.; Nikitenko, A.Y. Morphometric and molecular analyses for a new species Skrjabinolecithum pyriforme n. sp. (Digenea: Haploporidae) in mullet fish from the Primorsky Region, Russia. J. Helminthol. 2017, 91, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Besprozvannykh, V.V.; Atopkin, D.M.; Ngo, H.D.; Ha, N.V.; Tang, N.V.; Beloded, A.Y. Morphometric and molecular analyses of two digenean species from the mullet: Skrjabinolecithum spinosum n. sp. from the Russian southern Far East and Unisaccus tonkini n. sp. from Vietnam. J. Helminthol. 2018, 92, 713–724. [Google Scholar] [CrossRef]
- Andres, M.J.; Pulis, E.E.; Overstreet, R.M. Description of three species of Isorchis (Digenea: Atractotrematidae) from Australia. Acta Parasitol. 2016, 61, 590–601. [Google Scholar] [CrossRef] [Green Version]
- Atopkin, D.M.; Besprozvannykh, V.V.; Ha, D.N.; Nguyen, V.H.; Nguyen, V.T.; Chalenko, K.P. A new subfamily, Pseudohaploporinae subfam. n. (Digenea: Haploporidae), with morphometric and molecular analyses of two new species: Pseudohaploporus vietnamensis n. g., sp. n. and Pseudohaploporus planilizum n. g., sp. n. from Vietnamese mullet. Parasitol. Int. 2018, 69, 17–24. [Google Scholar] [CrossRef]
- Atopkin, D.M.; Besprozvannykh, V.V.; Ha, D.N.; Nguyen, V.H.; Khamatova, A.Y.; Vainutis, K.S. Morphometric and molecular analyses of Carassotrema koreanum Park 1938 and Elonginurus mugilus Lu, 1995 (Digenea: Haploporidae) Srivastava, 1937 from the Russian Far East and Vietnam. Parasitol. Res. 2019, 118, 2129–2137. [Google Scholar] [CrossRef]
- Atopkin, D.M.; Besprozvannykh, V.V.; Ha, D.N.; Nguyen, V.H.; Nguyen, V.T. New species and new genus of Pseudohaploporinae (Digenea): Pseudohaploporus pusitestis sp. n. and Parahaploporus elegantus n. g., sp. n. (Digenea: Pseudohaploporinae) from Vietnamese mullet fish. Parasitol. Int. 2020, 75, 102023. [Google Scholar] [CrossRef]
- Atopkin, D.M.; Besprozvannykh, V.V.; Ha, D.N.; Nguyen, V.H.; Nguyen, V.T. New species of Parasaccocoelium (Haploporidae) and new genus Pseudohaplosplanchnus (Haplosplanchnidae) from mullet fish in the Far East of Russia and Vietnam: Morphological and molecular data. J. Helminthol. 2020, 94, e154. [Google Scholar] [CrossRef]
- Truett, G.E. Preparation of genomic DNA from animal tissues. In The DNA Book: Protocols and Procedures for the Modern Molecular Biology; Kieleczawa, J., Ed.; Jones & Bartlett Publisher: Burlington, MA, USA, 2006; pp. 33–46. [Google Scholar]
- Matejusova, I.; Cunningham, C.O. The first complete monogenean ribosomal RNA gene operon: Sequence and secondary structure of the Gyrodactylus salaris Malmberg, 1957, large subunit ribosomal RNA gene. J. Parasitol. 2004, 90, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Tkach, V.V.; Littlewood, D.T.J.; Olson, P.D.; Kinsella, J.M.; Swiderski, Z. Molecular phylogenetic analysis of the Microphalloidea Ward, 1901 (Trematoda: Digenea). Syst. Parasitol. 2003, 56, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Besprozvannykh, V.V.; Tatonova, Y.V.; Shumenko, P.G. Life cycle, morphology of developmental stages of Metorchis ussuriensis sp. nov. (Trematoda: Opisthorchiidae), and phylogenetic relationships of other opisthorchiids. J. Zool. Syst. Evol. Res. 2019, 57, 24–40. [Google Scholar] [CrossRef] [Green Version]
- Luton, K.; Walker, D.; Blair, D. Comparisons of ribosomal internal transcribed spacers from two congeneric species of flukes (Platyhelminthes: Trematoda: Digenea). Mol. Biochem. Parasitol. 1992, 56, 323–327. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef] [Green Version]
- Posada, D. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr. Protoc. Bioinform. 2003, 6, 6.5.1–6.5.14. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModeltest2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Atopkin, D.M.; Beloded, A.Y.; Ngo, H.D. Molecular genetic characterization of the far eastern trematode Skrjabinolecithum spasskii Belous, 1954, (Digenea, Haploporidae), a parasite of mullets. Russ. J. Mol. Biol. 2015, 49, 422–429. [Google Scholar] [CrossRef]
- Atopkin, D.M.; Semenchenko, A.A.; Solodovnik, D.A.; Ivashko, Y.I.; Vinnikov, K.A. First next-generation sequencing data for Haploporidae (Digenea: Haploporata): Characterization of complete mitochondrial genome and ribosomal operon for Parasaccocoelium mugili Zhukov, 1971. Parasitol. Res. 2021, 120, 2037–2046. [Google Scholar] [CrossRef]
- Blasco-Costa, I.; Balbuena, J.A.; Kostadinova, A.; Olson, P.D. Interrelationships of the Haploporinae (Digenea: Haploporidae): A molecular test of the taxonomic framework based on morphology. Parasitol. Int. 2009, 58, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Andrade-Gómez, L.; García-Valera, M. Unexpected morphological and molecular diversity of trematode (Haploporidae: Forticulcitinae) parasites of mullets from the ocean Pacific coasts in Middle America. Parasitol. Res. 2021, 120, 55–72. [Google Scholar] [CrossRef]
- Andrade-Gómez, L.; González-García, M.T.; García-Valera, M. Phylogenetic affinities of Forticulcitinae (Haploporidae) parasites of mullet from the Americas, with the description of three new species and notes on the genera and key species. Syst. Parasitol. 2021, 98, 455–476. [Google Scholar] [CrossRef]
- Andres, M.J.; Curran, S.S.; Fayton, T.J.; Pulis, E.E.; Overstreet, R.M. An additional genus and two additional species of Forticulcitinae (Digenea: Haploporidae). Folia Parasitol. 2015, 62, 025. [Google Scholar] [CrossRef] [PubMed]
- Curran, S.S.; Tkach, V.V.; Overstreet, R.M. A review of Polylekithum Arnold, 1934 and its familial affinities using morphological and molecular data, with description of Polylekithum catahoulensis sp. nov. Acta Parasitologica 2006, 51, 238–248. [Google Scholar] [CrossRef]
- Curran, S.S.; Pulis, E.E.; Andres, M.J.; Overstreet, R.M. Two new species of Saccocoelioides (Digenea: Haploporidae) with phylogenetic analysis of the family, including species of Saccocoelioides from North, Middle and south America. J. Parasitol. 2018, 104, 221–239. [Google Scholar] [CrossRef]
- Pulis, E.; Fayton, T.; Curran, S.; Overstreet, R. A new species of Intromugil (Digenea: Haploporidae) and redescription of Intromugil mugilicolus. J. Parasitol. 2013, 99, 501–508. [Google Scholar] [CrossRef]
- Andres, M.J.; Pulis, E.E.; Curran, S.S.; Overstreet, R.M. On the systematics of some marine haploporids (Trematoda) with the description of a new species of Megasolena Linton, 1910. Parasitol. Int. 2018, 67, 805–815. [Google Scholar] [CrossRef]
- Huston, D.C.; Cutmore, S.C.; Cribb, T.H. Isorchis cannoni n. sp. (Digenea: Atroctotrematidae) from Great Barrier Reef rabbitfishes and the molecular elucidation of its life cycle. J. Helmointhology 2018, 92, 604–611. [Google Scholar] [CrossRef]
- Briscoe, A.G.; Bray, R.A.; Brabec, J.; Littlewood, D.T. The mitochondrial genome and ribosomal operon of Brachycladium goliath (Digenea: Brachycladiidae) recovered from a stranded minke whale. Parasitol. Int. 2016, 65, 271–275. [Google Scholar] [CrossRef]
- Martin, W.E. A new genus and species of haploporid trematode (Haploporidae: Trematoda) from Australian mullet. Bull. South. Calif. Acad. Sci. 1973, 72, 166–168. [Google Scholar]
- Martin, W.E. A new subfamily, two new genera, and three new species of haploporid trematodes. Proc. Helminthol. Soc. 1973, 40, 112–117. [Google Scholar]
- Zhukov, E.V. New genera of trematodes from marine fishes of India. Parazitologya 1972, 6, 346–350. [Google Scholar]
- Andrade-Gómez, L.; Pinacho-Pinacho, C.D.; Hernández-Orts, J.S.; Sereno-Uribe, A.L.; García-Valera, M. Morphological and molecular analyses of a new species of Saccocoelioidea Szidat, 1954 (Haploporidae Nicoll, 1914) in the fat sleeper Dominator maculates (Blosh) (Perciformes: Eleotridae) from the Gulf of Mexico. J. Helminthol. 2017, 91, 504–516. [Google Scholar] [CrossRef]



| Species | N | Definitive Host | Authors | Accession Number in the NCBI | |
|---|---|---|---|---|---|
| 28S | ITS1-5.8S-ITS2 | ||||
| Haploporidae | |||||
| Waretrematinae | |||||
| Paraunisaccoides elegans n. sp. | 4/2 | Planiliza subviridis | Present study | KY501639–KY501641 | KY501642–KY501644 |
| Unisaccus halongi n. sp. | 3/3 | Crenimugil seheli | Present study | OK644190-OK644192 | OK644196-OK644198 |
| Unisaccus tonkini | 1/1 | Moolgarda cunnesius | [6] | MF176843 | MF176838 |
| Skrjabinolecithum pyriforme | 1/1 | Planiliza haematocheila | [5] | HE806359 | LN864990 |
| Skrjabinolecithum spinosum | 1/1 | Planiliza haematocheila | [6] | MF176834 | MF176831 |
| Skrjabinolecithum. spasskii | 3/3 | Planiliza haematocheila | [21] | LN614538, HG530210, HG530207 | LK022754, HE806371, HG530228 |
| Parasaccocoelium mugili | 1/1 | Planiliza haematocheila | [22] | MW813991 | |
| Parasaccocoelium armatum | -/2 | Mugil cephalus | [11] | - | MT298950–MT298951 |
| Parasaccocoelium haematochelium | -/2 | Liza haematocheila | [4] | - | HF548466–HF548467 |
| Parasaccocoelium polyovum | -/2 | Liza haematocheila | [4] | - | HF548477–HF548478 |
| Elonginurus mugilus | 1/1 | Mugil cephalus | [9] | MH763766 | MH763761 |
| Carassotrema koreanum | 3/3 | Carassius gibelio | [9] | MH763763–MH763765 | MH763758–MH763760 |
| Carassotrema sp. | 1/1 | unpublished | MH285255 | ||
| Spiritestis herveyensis | 1/1 | Moolgarda seheli | [2] | KC206500 | |
| Capitimitta costata | 1/1 | Selenotoca multifasciata | [2] | KC206497 | |
| Capitimitta darwinensis | 1/1 | Selenotoca multifasciata | [2] | KC206498 | |
| Pseudohaploporinae | |||||
| Parahaploporus elegantus | 10 | Moolgarda seheli | [10] | MN639712-MN639721 | |
| Pseudohaploporus vietnamensis | 6/8 | Osteomugil engeli | [8] | MF774420–MF774421, MF774423–MF774426 | MF774427–MF774429; MF774436–MF774440 |
| Pseudohaploporus vietnamensis | 1/1 | Moolgarda seheli | [8] | MF774422 | MF774431 |
| Pseudohaploporus planilizum | 3/3 | Planiliza subviridis | [8] | MF774417–MF774419 | MF774433–MF774435 |
| Pseudohaploporus pusitestis | 2/2 | Moolgarda seheli | [10] | MH986037, MH986038 | MF774430, MF774432 |
| Haploporinae | |||||
| Saccocoelium brayi | 1/1 | Liza saliens | [23] | FJ211234 | FJ211244 |
| Saccocoelium cephali | 1/1 | Mugil cephalus | [23] | FJ211233 | FJ211243 |
| Saccocoelium obesum | 2/2 | Liza ramada | [23] | FJ211259–FJ211260 | FJ211265–FJ211266 |
| Saccocoelium tensum | 2/2 | Liza ramada | [23] | FJ211257–FJ211258 | FJ211263–FJ211264 |
| Dicrogaster contracta | 1/1 | Liza aurata | [23] | FJ211261 | FJ211267 |
| Dicrogaster perpusilla | 1/1 | Liza ramada | [23] | FJ211238 | FJ211248 |
| Lecithobotrys putrescen | 1/1 | Liza saliens | [23] | FJ211236 | FJ211246 |
| Litosaccus_brisbanensis | 1/1 | Mugil cephalus | [3] | KM253765 | |
| Haploporus benedeni | 1/1 | Liza ramado | [23] | FJ211237 | FJ211247 |
| Ragaia lizae | 1/1 | Liza aurata | [23] | FJ211235 | FJ211245 |
| Forticulcitinae | |||||
| Forticulcita isabelae | 1/1 | Mugil curema | [24] | MT957787 | MT957640 |
| Forticulcita macropharyngis | 1/1 | Mugil curema | [25] | MW796513 | MW796548 |
| Forticulcita minuta | 1/1 | Mugil cephalus | [24] | MT957822 | MT957660 |
| Forticulcita venezuelensis | 1/1 | Mugil curema | [25] | MW796522 | MW796549 |
| Forticulcita gibsoni | 1/1 | Mugil cephalus | [24] | FJ211239 | FJ211249 |
| F. apiensis | 1/1 | Mugil cephalus | [26] | KP761087 | |
| F. platana | 1/1 | Mugil liza | [26] | KP761086 | |
| Xiha fastigata | 1/1 | Mugil cephalus | [26] | KP761088 | |
| Overstreetoides_pacificus | 2/2 | Mugil curema | [24] | MT957753–MT957754 | MT957629–MT957630 |
| Ekuarhuni papillatum | 2/2 | Mugil sp. | [24] | MT957687–MT957688 | MT957587–MT957588 |
| Ekuarhuni mexicanum | 2/2 | Mugil sp. | [24] | MW796487–MW796488 | MW796524–MW796525 |
| Chalcinotrematinae | |||||
| Saccocoelioides sp. | 1/1 | Unidentified molly (Poecilidae) | [27] | EF032696 | - |
| Saccocoelioides beauforti | 1/1 | Mugil cephalus | [28] | MG925104 | MG925103 |
| Saccocoelioides elongatus | 1/1 | Prochilodus lineatus | [28] | MG925108 | MG925107 |
| Saccocoelioides magnus | 1/1 | Cyphocharax voga | [28] | MG925112 | MG925111 |
| Saccocoelioides nanii | 1/1 | Prochilodus lineatus | [28] | MG925114 | MG925113 |
| Saccocoelioides orosiensis | 1/1 | Poecilia gillii | [28] | MG925118 | MG925117 |
| Saccocoelioides tkachi | 1/1 | Astyanax aeneus | [28] | MG925122 | MG925121 |
| Intromugil mugilicolus | 1/1 | Mugil cephalus | [29] | KC430096 | |
| Intromugil alachuaensis | 1/1 | Mugil cephalus | [29] | KC430095 | |
| Hapladeninae | |||||
| Hapladena acanthuri | 1/1 | Acanthurus chirurgus | [30] | MH244119 | |
| Hapladena cf. varia | 1/1 | Acanthurus chirurgus | [30] | MH244120 | |
| Megasoleninae | |||||
| Megasolena hysterospina | 1/1 | Archosargus rhomboidalis | [30] | MH244121 | |
| Megasolena sp. m MA-2018 | 1/1 | Holacanthus ciliaris | [30] | MH244122 | |
| Cadenatelinae | |||||
| Cadenatella americana | 1/1 | Kyphosus sectatrix | [30] | MH244117 | |
| Cadenatella floridae | 1/1 | Kyphosus incisor | [30] | MH244118 | |
| Atractotrematidae | |||||
| Isorchis anomalus | 1/1 | Chanos chanos | [7] | KU873018 | |
| Isorchis currani | 1/1 | Selenotoca multifasciata | [31] | KU873017 | |
| Isorchis megas | 1/1 | Selenotoca multifasciata | [7] | KU873015 | |
| Brachycladiidae | |||||
| Brachycladium goliath | 1/1 | Balaenoptera acutorostrata | [32] | KR703279 | |
| Monorchiidae | |||||
| Hurleytrematoides chaetodoni | 1/1 | Chaetodon striatus | [30] | MH244116 | |
| Paraunisaccoides elegans n. sp. | Skrjabinolecithumlobolecitum (Martin, 1973) | S. vitellosum (Martin, 1973) | Unisacus halongi n. sp. | S. indicum (Zhukov, 1972) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Holotype | Range | Mean | Holotype | Range | Mean | ||||
| Body length | 1217 | 1124–1232 | 1170 | 1850; 2070 | 510–790 | 920 | 920–1390 | 1170 | 1000–1200 |
| Body width | 354 | 323–370 | 347 | 266; 406 | 140–300 | 420 | 390–500 | 430 | 280–370 |
| Body length/width% | 29.1 | 26.0–32.9 | 29.7 | - | 45.7 | 31.7–45.7 | 36.8 | - | |
| Forebody length | 501 | 424–508 | 469 | - | 0.347 | 347–396 | 364 | - | |
| Body/forebody length% | 41,2 | 37.5–44.0 | 40.1 | - | 32.2 | 28.2–37.7 | 31.1 | - | |
| Oral sucker length | 96 | 77–104 | 92 | 96; 112 | 50–80 | 96 | 92–127 | 104 | 83–120 |
| Oral sucker width | 96 | 92–112 | 100 | 100; 112 | 59–90 | 131 | 116–154 | 139 | 110–120 |
| Ventral sucker length | 123 | 112–139 | 125 | 93; 143 | 56–74 | 127 | 96–142 | 120 | 120–150 |
| Ventral sucker width | 131 | 127–135 | 129 | 78; 156 | 56–74 | 127 | 112–150 | 124 | 140–170 |
| Ventral/oral sucker length ratio | 1:1.28 | 1:1.17–1.71 | 1:1.36 | - | 1:1.38 | 1: 0.91–1.48 | 1:1.15 | - | |
| Ventral/oral sucker width ratio | 1:1.36 | 1:1.13–1.47 | 1:1.29 | - | 1:1.09 | 1: 0.73–1.09 | 1:1.06 | - | |
| Prepharynx length | 539 | 377–566 | 483 | 426; 684 | 90–109 | 227 | 227–354 | 306 | 120–170 |
| Pharynx length | 77 | 77–116 | 93 | 109; 112 | 44–80 | 77 | 77–116 | 95 | 110–140 |
| Pharynx width | 85 | 85–123 | 100 | 131; 137 | 59–110 | 89 | 89–135 | 117 | 80–110 |
| Oesophagus length | 15 | 0–154 | - | 112; 249 | 60 | 58 | 23–58 | 49 | - |
| Ovary length | 62 | 46–77 | 71 | 109; 140 | 31–62 | 69 | 69–116 | 96 | 62–83 |
| Ovary width | 62 | 54–77 | 64 | 100; 109 | 31–62 | 69 | 69–116 | 97 | 62–83 |
| Testis length | 177 | 162–219 | 206 | 239; 345 | 75–165 | 150 | 150–270 | 180 | 210–300 |
| Testis width | 193 | 154–235 | 198 | 168; 202 | 50–90 | 193 | 139–227 | 184 | 120–140 |
| Hermaphroditic sac length | 254 | 181–270 | 220 | 258; 286 | 90 | 177 | 154–231 | 186 | - |
| Hermaphroditic sac width | 96 | 100–123 | 114 | 96; 118 | 50 | 104 | 104–173 | 138 | - |
| Eggs length | 62–65 | 62–65 | - | 71–93 | 59–65 | 62–73 | 62–73 | - | 71–79 |
| Eggs width | 50–54 | 50–54 | - | 56–59 | 42 | 42–50 | 42–50 | - | 39–43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atopkin, D.M.; Besprozvannykh, V.V.; Beloded, A.Y.; Ha, N.D.; Nguyen, H.V.; Nguyen, T.V. Restoration of the Genus Paraunisaccoides Martin, 1973 (Digenea: Haploporidae) and Description of P. elegans n. sp. and Unisaccus halongi n. sp. from Mugilid Fish in Vietnam. Diversity 2022, 14, 639. https://0-doi-org.brum.beds.ac.uk/10.3390/d14080639
Atopkin DM, Besprozvannykh VV, Beloded AY, Ha ND, Nguyen HV, Nguyen TV. Restoration of the Genus Paraunisaccoides Martin, 1973 (Digenea: Haploporidae) and Description of P. elegans n. sp. and Unisaccus halongi n. sp. from Mugilid Fish in Vietnam. Diversity. 2022; 14(8):639. https://0-doi-org.brum.beds.ac.uk/10.3390/d14080639
Chicago/Turabian StyleAtopkin, D. M., V. V. Besprozvannykh, A. Yu. Beloded, N. D. Ha, H. V. Nguyen, and T. V. Nguyen. 2022. "Restoration of the Genus Paraunisaccoides Martin, 1973 (Digenea: Haploporidae) and Description of P. elegans n. sp. and Unisaccus halongi n. sp. from Mugilid Fish in Vietnam" Diversity 14, no. 8: 639. https://0-doi-org.brum.beds.ac.uk/10.3390/d14080639