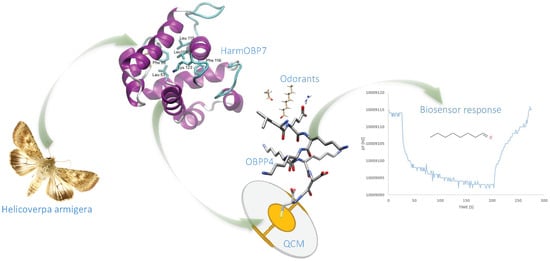
A Highly Selective Biosensor Based on Peptide Directly Derived from the HarmOBP7 Aldehyde Binding Site
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Modelling
2.2. Peptide Synthesis and Deposition
2.3. Gas Mixtures and Measurements Setup
3. Results and Discussion
3.1. Peptide-Based Sensor Film Characterization
3.2. Virtual Docking
3.3. OBPPs-Based Biosensor Parameters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wasilewski, T.; Gębicki, J.; Kamysz, W. Bioelectronic nose: Current status and perspective. Biosens. Bioelectron. 2017, 87, 480–494. [Google Scholar] [CrossRef] [PubMed]
- McGann, J.P. Poor human olfaction is a 19th-century myth. Science 2017, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasilewski, T.; Gębicki, J.; Kamysz, W. Advances in olfaction-inspired biomaterials applied to bioelectronic noses. Sens. Actuators B Chem. 2018, 257, 511–537. [Google Scholar] [CrossRef]
- Wasilewski, T.; Szulczyński, B.; Kamysz, W.; Gębicki, J.; Namieśnik, J. Evaluation of three peptide immobilization techniques on a qcm surface related to acetaldehyde responses in the gas phase. Sensors 2018, 18, 3942. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Deng, L.; Zhu, X.-M.; Fan, Y.; Hu, J.-N.; Li, J.; Deng, Z.-Y. Novel Approach To Evaluate the Oxidation State of Vegetable Oils Using Characteristic Oxidation Indicators. J. Agric. Food Chem. 2014, 62, 12545–12552. [Google Scholar] [CrossRef] [PubMed]
- Zepka, L.Q.; Wagner, R.; Jacob-Lopes, E.; Daltoé, M.M.; Santos, A.B.; Torri, A.F.; Donadel, J.Z.; Queiroz, M.I. Study of the Volatile Compounds Useful for the Characterization of Frozen Anchoita (Engraulis anchoita) by SPME-GC-MS. In Flavour Science; Elsevier: Amsterdam, The Netherlands, 2014; pp. 169–172. [Google Scholar] [CrossRef]
- Fuchs, P.; Loeseken, C.; Schubert, J.K.; Miekisch, W. Breath gas aldehydes as biomarkers of lung cancer. Int. J. Cancer 2010, 126, 2663–2670. [Google Scholar] [CrossRef]
- Sankaran, S.; Khot, L.R.; Panigrahi, S. Biology and applications of olfactory sensing system: A review. Sens. Actuators B Chem. 2012, 171–172, 1–17. [Google Scholar] [CrossRef]
- Valle, M. Bioinspired sensor systems. Sensors 2011, 11, 10180–10186. [Google Scholar] [CrossRef]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef]
- Khadka, R.; Aydemir, N.; Carraher, C.; Hamiaux, C.; Colbert, D. An ultrasensitive electrochemical impedance-based biosensor using insect odorant receptors to detect odorants. Biosens. Bioelectron. 2019, 126, 207–213. [Google Scholar] [CrossRef]
- Hurot, C.; Brenet, S.; Buhot, A.; Barou, E.; Belloir, C.; Briand, L. Highly sensitive olfactory biosensors for the detection of volatile organic compounds by surface plasmon resonance imaging. Biosens. Bioelectron. 2019, 123, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Kotlowski, C.; Larisika, M.; Guerin, P.M.; Kleber, C.; Kröber, T.; Mastrogiacomo, R.; Nowak, C.; Pelosi, P.; Schütz, S.; Schwaighofer, A.; et al. Chemical Fine discrimination of volatile compounds by graphene-immobilized odorant-binding proteins. Sens. Actuators B Chem. 2018, 256, 564–572. [Google Scholar] [CrossRef]
- Boyle, S.M.; McInally, S.; Ray, A. Expanding the olfactory code by in silico decoding of odor-receptor chemical space. Elife 2013. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Zhu, J.; Knoll, W. Odorant-Binding Proteins as Sensing Elements for Odour Monitoring. Sensors 2018, 18, 3248. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, A.J.M.; Oliveira, A.R.; Roque, A.C.A. Protein- and Peptide-Based Biosensors in Artificial Olfaction. Trends Biotechnol. 2018, 36, 1244–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Man, O.; Gilad, Y.; Lancet, D. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 2004, 13, 240–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, C.; Miyake, J. Chapter 8. Combinatorially Developed Peptide Receptors for Biosensors. In Combinatorial Methods for Chemical and Biological Sensors, GE Global; Potyrailo, A.R., Ed.; Springer: Niskayuna, NY, USA, 2016. [Google Scholar]
- Boon, C.L.; Frost, D.; Chakrabartty, A. Identification of stable helical bundles from a combinatiorial library of amphipathic peptides. Biopolym. Pept. Sci. Sect. 2004, 76, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.C.; Gaj, T.; Ghosh, I. Highly selective cyclic peptide ligands for neutravidin and avidin identified by phage display. Chem. Biol. Drug Des. 2006, 68, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, J.; Boyd, B.J. Peptide-based biosensors. Talanta 2015, 136, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Ruotolo, B.T.; Verbeck, G.F.; Thomson, L.M.; Gillig, K.J.; Russell, D.H. Observation of conserved solution-phase secondary structure in gas-phase tryptic peptides. J. Am. Chem. Soc. 2002, 124, 4214–4215. [Google Scholar] [CrossRef]
- Farrar, D.; West, J.E.; Busch-Vishniac, I.J.; Yu, S.M. Fabrication of polypeptide-based piezoelectric composite polymer film. Scr. Mater. 2008, 59, 1051–1054. [Google Scholar] [CrossRef]
- Lu, H.-H.H.; Rao, Y.K.; Wu, T.-Z.Z.; Tzeng, Y.-M.M. Direct characterization and quantification of volatile organic compounds by piezoelectric module chips sensor. Sens. Actuators B Chem. 2009, 137, 741–746. [Google Scholar] [CrossRef]
- Diociaiuti, M.; Gaudiano, M.C.; Malchiodi-Albedi, F. The slowly aggregating salmon Calcitonin: A useful tool for the study of the amyloid oligomers structure and activity. Int. J. Mol. Sci. 2011, 12, 9277–9295. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, S.; Panigrahi, S.; Mallik, S. Odorant binding protein based biomimetic sensors for detection of alcohols associated with Salmonella contamination in packaged beef. Biosens. Bioelectron. 2011, 26, 3103–3109. [Google Scholar] [CrossRef] [PubMed]
- Son, M.; Kim, D.; Kang, J.; Lim, J.H.; Lee, S.H.; Ko, H.J.; Hong, S.; Park, T.H. Bioelectronic Nose Using Odorant Binding Protein-Derived Peptide and Carbon Nanotube Field-Effect Transistor for the Assessment of Salmonella Contamination in Food. Anal. Chem. 2016, 88, 11283–11287. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-Q.; Zhang, S.; Luo, J.-Y.; Cui, J.-J.; Ma, Y.; Dong, S.-L. Two Minus-C odorant binding proteins from Helicoverpa armigera display higher ligand binding affinity at acidic pH than neutral pH. J. Insect Physiol. 2013, 59, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-L.; Huang, L.-Q.; Pelosi, P.; Wang, C.-Z. A Lysine at the C-Terminus of an Odorant-Binding Protein is Involved in Binding Aldehyde Pheromone Components in Two Helicoverpa Species. PLoS ONE 2013, 8, e55132. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Hwang, J.-K.; Yang, J.-M. (PS)2-v2: Template-based protein structure prediction server. BMC Bioinform. 2009, 10, 366. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Genz, M.; Balke, K.; Bornscheuer, U.T. The effect of disulfide bond introduction and related Cys/Ser mutations on the stability of a cyclohexanone monooxygenase. J. Biotechnol. 2015, 214, 199–211. [Google Scholar] [CrossRef]
- Zhou, P.; Tian, F.; Lv, F.; Shang, Z. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins Struct. Funct. Bioinforma. 2009, 76, 151–163. [Google Scholar] [CrossRef]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef] [PubMed]
- Tuffery, P.; Thevenet, P.; Guyon, F.; Shen, Y.; Derreumaux, P.; Maupetit, J. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojciechowski, M. Simplified AutoDock force field for hydrated binding sites. J. Mol. Graph. Model. 2017, 78, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Srisombat, L.; Jamison, A.C.; Lee, T.R. Stability: A key issue for self-assembled monolayers on gold as thin-film coatings and nanoparticle protectants. Colloids Surfaces A Physicochem. Eng. Asp. 2011, 390, 1–19. [Google Scholar] [CrossRef]
- Latif, U.; Can, S.; Hayden, O.; Grillberger, P.; Dickert, F.L. Sauerbrey and anti-Sauerbrey behavioral studies in QCM sensors—Detection of bioanalytes. Sens. Actuators B Chem. 2013, 176, 825–830. [Google Scholar] [CrossRef]
- Anselmi, C.; Buonocore, A.; Centini, M.; Maffei, R.; Hatt, H. The human olfactory receptor 17-40: Requisites for fitting into the binding pocket. Comput. Biol. Chem. 2011, 35, 159–168. [Google Scholar] [CrossRef]
- Desimoni, E.; Brunetti, B. About Estimating the Limit of Detection by the Signal to Noise Approach. Pharm. Anal. Acta 2015, 6. [Google Scholar] [CrossRef]
- Wessa, T.; Göpel, W. Molecular recognition: Supramolecular, polymeric and biomimetic coatings for chemical sensors and chromatographic columns. Fresenius J. Anal. Chem. 1998, 361, 239–245. [Google Scholar] [CrossRef]
- di Natale, C.; Macagnano, A.; Davide, F.; D’Amico, A.; Paolesse, R.; Boschi, T.; Faccio, M.; Ferri, G. An electronic nose for food analysis. Sens. Actuators B Chem. 1997, 44, 521–526. [Google Scholar] [CrossRef]
- Compagnone, D.; Fusella, G.C.C.; del Carlo, M.; Pittia, P.; Martinelli, E.; Tortora, L.; Paolesse, R.; di Natale, C.; Pittia, P.; Paolesse, R.; et al. Gold nanoparticles-peptide based gas sensor arrays for the detection of food aromas. Biosens. Bioelectron. 2013, 42, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.J.; Park, T.H. Piezoelectric olfactory biosensor: Ligand specificity and dose-dependence of an olfactory receptor expressed in a heterologous cell system. Biosens. Bioelectron. 2005, 20, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Hun, S.; Beom, S.; Jin, H.; June, S.; Hyun, T.; Lee, S.H.; Jun, S.B.; Ko, H.J.; Kim, S.J.; Park, T.H. Cell-based olfactory biosensor using microfabricated planar electrode. Biosens. Bioelectron. 2009, 24, 2659–2664. [Google Scholar] [CrossRef]




| Compound | OBPP1 [kcal/mol] | OBPP2 [kcal/mol] | OBPP3 [kcal/mol] | OBPP4 [kcal/mol] |
|---|---|---|---|---|
| Octanal | −5.03 | −5.33 | −5.03 | −5.28 |
| Acetaldehyde | −2.41 | −2.40 | −2.43 | −2.33 |
| Benzaldehyde | −4.95 | −4.59 | −4.19 | −4.56 |
| Ethanol | −2.55 | −2.48 | −2.48 | −2.37 |
| Acetone | −3.00 | −2.88 | −3.02 | −2.82 |
| Dimethyl Sulphide | −2.34 | −2.14 | −2.40 | −2.12 |
| Trimethyl Amine | −1.42 | −2.32 | −2.57 | −1.71 |
| Toluene | −4.47 | −4.17 | −3.57 | −3.99 |
| Ammonia | −1.23 | −1.74 | −1.53 | −1.01 |
| Compounds | Parameters | OBPP1 | OBPP2 | OBPP3 | OBPP4 |
|---|---|---|---|---|---|
| Ammonia | S·103 [Hz/ppm] | 0.7 | 4.9 | 0.1 | 0.5 |
| LOD [ppm v/v] | >1500 | >1500 | >1500 | >1500 | |
| R2 | 0.92 | 0.95 | 0.313 | 0.661 | |
| Acetone | S·103 [Hz/ppm] | 16.3 | 2.6 | 3.3 | 4.0 |
| LOD | >1500 | >1500 | >1500 | >1500 | |
| R2 | 0.939 | 0.818 | 0.801 | 0.771 | |
| Dimethyl sulphide | S·103 [Hz/ppm] | 2.0 | 9.7 | no response | no response |
| LOD | >1500 | >1500 | no response | no response | |
| R2 | 0.458 | 0.951 | no response | no response | |
| Acetaldehyde | S·103 [Hz/ppm] | 116.8 | 142.8 | 29.1 | 27.5 |
| LOD | 243.4 | 574.0 | 571.1 | >1500 | |
| R2 | 0.998 | 0.987 | 0.987 | 0.987 | |
| Ethanol | S·103 [Hz/ppm] | 2.3 | 13.5 | no response | no response |
| LOD | >1500 | 1224.0 | no response | no response | |
| R2 | 0.751 | 0.939 | no response | no response | |
| Trimethylamine | S·103 [Hz/ppm] | 20.9 | 53.7 | 145.6 | 159.6 |
| LOD | >1500 | 424.5 | 175.5 | 105.1 | |
| R2 | 0.96 | 0.957 | 0.992 | 0.997 | |
| Benzaldehyde | S·103 [Hz/ppm] | 91.5 | 55.8 | 26.1 | 25.0 |
| LOD | 678.6 | 650.3 | 892.0 | 485.3 | |
| R2 | 0.943 | 0.947 | 0.905 | 0.970 | |
| Toluene | S·103 [Hz/ppm] | 63.3 | 31.3 | no response | no response |
| LOD | 906.2 | 826.3 | no response | no response | |
| R2 | 0.895 | 0.911 | no response | no response | |
| Octanal | S·103 [Hz/ppm] | 16.4 | 67.7 | 335.4 | 667.5 |
| LOD | 455.6 | 293.9 | 48.5 | 37.5 | |
| R2 | 0.940 | 0.974 | 0.999 | 0.997 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wasilewski, T.; Szulczyński, B.; Wojciechowski, M.; Kamysz, W.; Gębicki, J. A Highly Selective Biosensor Based on Peptide Directly Derived from the HarmOBP7 Aldehyde Binding Site. Sensors 2019, 19, 4284. https://0-doi-org.brum.beds.ac.uk/10.3390/s19194284
Wasilewski T, Szulczyński B, Wojciechowski M, Kamysz W, Gębicki J. A Highly Selective Biosensor Based on Peptide Directly Derived from the HarmOBP7 Aldehyde Binding Site. Sensors. 2019; 19(19):4284. https://0-doi-org.brum.beds.ac.uk/10.3390/s19194284
Chicago/Turabian StyleWasilewski, Tomasz, Bartosz Szulczyński, Marek Wojciechowski, Wojciech Kamysz, and Jacek Gębicki. 2019. "A Highly Selective Biosensor Based on Peptide Directly Derived from the HarmOBP7 Aldehyde Binding Site" Sensors 19, no. 19: 4284. https://0-doi-org.brum.beds.ac.uk/10.3390/s19194284