Estimation of Leaf Nitrogen Content in Wheat Based on Fusion of Spectral Features and Deep Features from Near Infrared Hyperspectral Imagery
Abstract
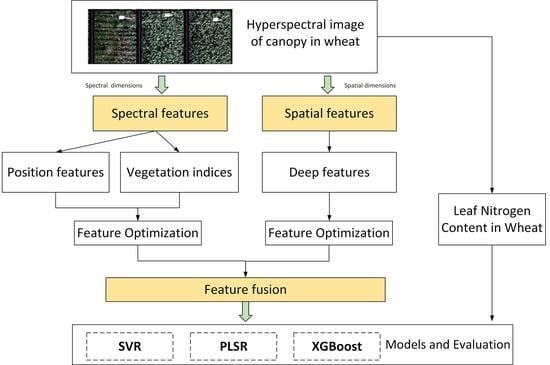
:1. Introduction
2. Data and Methods
2.1. Study Site and Experimental Design
2.2. Data Collection
2.3. Spectral Features and Deep Features
2.3.1. Vegetation Indices
2.3.2. Position Features
2.3.3. Deep Features
2.4. Feature Optimization Method
2.4.1. Random Forest Algorithm
2.4.2. Pearson Correlation Coefficient Method
2.5. Regression Method
2.5.1. Partial Least Squares Regression
2.5.2. Support Vector Regression
2.5.3. Gradient Boosting Decision Tree
3. Results and Analysis
3.1. Optimization of Vegetation Indices
3.2. Optimization of Position Features
3.3. Optimization of Deep Features
3.4. Comparison of Models for Estimating LNC in Winter
4. Discussion
4.1. Deep Features and Spectral Features
4.2. The Necessity of Extracting Deep Features from Hyperspectral Images
4.3. Different Models and Different Features
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, Y.; Tian, Y.; Yao, X.; Liu, X.; Cao, W. Analysis of Common Canopy Reflectance Spectra for Indicating Leaf Nitrogen Concentrations in Wheat and Rice. Plant Prod. Sci. 2007, 10, 400–411. [Google Scholar] [CrossRef]
- Yang, B.; Wang, M.; Sha, Z.; Wang, B.; Chen, J.; Yao, X.; Cheng, T.; Cao, W.; Zhu, Y. Evaluation of Aboveground Nitrogen Content of Winter Wheat Using Digital Imagery of Unmanned Aerial Vehicles. Sensors 2019, 19, 4416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabatel, G.; Al Makdessi, N.; Ecarnot, M.; Roumet, P. A spectral correction method for multi-scattering effects in close range hyperspectral imagery of vegetation scenes: Application to nitrogen content assessment in wheat. Adv. Anim. Biosci. 2017, 8, 353–358. [Google Scholar] [CrossRef]
- He, L.; Zhang, H.-Y.; Zhang, Y.-S.; Song, X.; Feng, W.; Kang, G.-Z.; Wang, C.-Y.; Guo, T.-C. Estimating canopy leaf nitrogen concentration in winter wheat based on multi-angular hyperspectral remote sensing. Eur. J. Agron. 2016, 73, 170–185. [Google Scholar] [CrossRef]
- Vigneau, N.; Ecarnot, M.; Rabatel, G.; Roumet, P. Potential of field hyperspectral imaging as a non destructive method to assess leaf nitrogen content in Wheat. Field Crop. Res. 2011, 122, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Song, X.; Li, Z.; Yang, G.; Guo, W.; Tan, C.; Chen, L. Estimation of winter wheat nitrogen nutrition index using hyperspectral remote sensing. Trans. Chin. Soc. Agric. Eng. 2014, 30, 191–198. [Google Scholar]
- Liu, H.; Zhu, H.; Li, Z.; Yang, G. Quantitative analysis and hyperspectral remote sensing of the nitrogen nutrition index in winter wheat. Int. J. Remote. Sens. 2019, 41, 858–881. [Google Scholar] [CrossRef]
- Feng, W.; Zhang, H.; Zhang, Y.; Qi, S.; Heng, Y.; Guo, B.; Ma, D.; Guo, T. Remote detection of canopy leaf nitrogen concen-tration in winter wheat by using water resistance vegetation indices from in-situ hyperspectral data. Field Crop. Res. 2016, 198, 238–246. [Google Scholar] [CrossRef]
- Ye, M.; Ji, C.; Chen, H.; Lei, L.; Qian, Y. Residual deep PCA-based feature extraction for hyperspectral image classifica-tion. Neural Comput. Appl. 2020, 32, 14287–14300. [Google Scholar] [CrossRef]
- Uddin, M.P.; Al Mamun, M.; Hossain, M.A. Effective feature extraction through segmentation-based folded-pca for hyper-spectral image classification. Int. J. Remote Sens. 2019, 40, 7190–7220. [Google Scholar] [CrossRef]
- Li, Y.; Ge, C.; Sun, W.; Peng, J.; Du, Q.; Wang, K. Hyperspectral and lidar data fusion classification using superpixel segmen-tation-based local pixel neighborhood preserving embedding. Remote Sens. 2019, 11, 550. [Google Scholar] [CrossRef] [Green Version]
- Bandos, T.V.; Bruzzone, L.; Camps-Valls, G. Classification of Hyperspectral Images With Regularized Linear Discriminant Analysis. IEEE Trans. Geosci. Remote. Sens. 2009, 47, 862–873. [Google Scholar] [CrossRef]
- Leemans, V.; Marlier, G.; Destain, M.-F.; Dumont, B.; Mercatoris, B. Estimation of leaf nitrogen concentration on winter wheat by multispectral imaging. Hyperspectral Imaging Sens. Innov. Appl. Sens. Stand. 2017, 102130I. [Google Scholar] [CrossRef] [Green Version]
- Mutanga, O.; Skidmore, A.K. Hyperspectral band depth analysis for a better estimation of grass biomass (Cenchrus ciliaris) measured under controlled laboratory conditions. Int. J. Appl. Earth Obs. Geoinf. 2004, 5, 87–96. [Google Scholar] [CrossRef]
- Ghasemzadeh, A.; Demirel, H. 3D discrete wavelet transform-based feature extraction for hyperspectral face recognition. IET Biom. 2018, 7, 49–55. [Google Scholar] [CrossRef]
- Cao, X.; Xu, L.; Meng, D.; Zhao, Q.; Xu, Z. Integration of 3-dimensional discrete wavelet transform and Markova random field for hyperspectral image classification. Neurocomputing 2017, 226, 90–100. [Google Scholar] [CrossRef]
- Li, H.-C.; Zhou, H.; Pan, L.; Du, Q. Gabor feature-based composite kernel method for hyperspectral image classification. Electron. Lett. 2018, 54, 628–630. [Google Scholar] [CrossRef]
- Jia, S.; Shen, L.; Li, Q. Gabor Feature-Based Collaborative Representation for Hyperspectral Imagery Classification. IEEE Trans. Geosci. Remote. Sens. 2015, 53, 1118–1129. [Google Scholar] [CrossRef]
- Li, W.; Chen, C.; Su, H.; Du, Q. Local binary patterns and extreme learning machine for hyperspectral imagery classifica-tion. IEEE Trans. Geoence Remote Sens. 2015, 53, 3681–3693. [Google Scholar] [CrossRef]
- Deng, Z.P.; Sun, H.; Zhou, S.L.; Zhao, J.P.; Lei, L.; Zou, H.X. Multi-scale object detection in remote sensing imagery with convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2018, 145, 3–22. [Google Scholar] [CrossRef]
- Zheng, H.; Li, W.; Jiang, J.; Liu, Y.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; Zhang, Y.; Yao, X. A Comparative Assessment of Different Modeling Algorithms for Estimating Leaf Nitrogen Content in Winter Wheat Using Multispectral Images from an Unmanned Aerial Vehicle. Remote. Sens. 2018, 10, 2026. [Google Scholar] [CrossRef] [Green Version]
- Alam, F.I.; Zhou, J.; Liew, W.C.; Jia, X.; Chanussot, J.; Gao, Y. Conditional random field and deep feature learning for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1612–1628. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep Feature Extraction and Classification of Hyperspectral Images Based on Convolutional Neural Networks. IEEE Trans. Geosci. Remote. Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Yu, X.; Zhang, P.; Yu, A.; Fu, Q.; Wei, X. Supervised Deep Feature Extraction for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote. Sens. 2017, 56, 1909–1921. [Google Scholar] [CrossRef]
- Pan, B.; Shi, Z.W.; Xu, X. MugNet: Deep learning for hyperspectral image classification using limited samples. ISPRS J. Photogramm. Remote Sens. 2018, 145, 108–119. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G.; Gu, Y. Deep Learning-Based Classification of Hyperspectral Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Xu, S.; Sun, X.; Lu, H.; Zhang, Q. Detection of Type, Blended Ratio, and Mixed Ratio of Pu’er Tea by Using Electronic Nose and Visible/Near Infrared Spectrometer. Sensors 2019, 19, 2359. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Gao, Y.; Yan, Q.; Qi, L.; Zhu, Y.; Wang, B. Estimation Method of Soluble Solid Content in Peach Based on Deep Features of Hyperspectral Imagery. Sensors 2020, 20, 5021. [Google Scholar] [CrossRef]
- Hasan, M.; Chopin, J.P.; Laga, H.; Miklavcic, S.J. Detection and analysis of wheat spikes using Convolutional Neural Networks. Plant Methods 2018, 14, 100. [Google Scholar] [CrossRef] [Green Version]
- Condori, R.H.M.; Romualdo, L.M.; Bruno, O.M.; de Cerqueira Luz, P.H. Comparison Between Traditional Texture Methods and Deep Learning Descriptors for Detection of Nitrogen Deficiency in Maize Crops. In Proceedings of the 2017 Workshop of Computer Vision (WVC), Natal, Brazil, 30 October–1 November 2017; pp. 7–12. [Google Scholar]
- Moghimi, A.; Yang, C.; Anderson, J.A. Aerial hyperspectral imagery and deep neural networks for high-throughput yield phenotyping in wheat. Comput. Electron. Agric. 2020, 172, 105299. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Luo, X.; Jin, J.; Wang, L.; Zhang, L.; Liu, J.; Zhang, Z. Improving High-Throughput Phenotyping Using Fusion of Close-Range Hyperspectral Camera and Low-Cost Depth Sensor. Sensors 2018, 18, 2711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, K.; Deng, X.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y.; Ustin, S.L.; Cheng, T. Assessing the Spectral Properties of Sunlit and Shaded Components in Rice Canopies with Near-Ground Imaging Spectroscopy Data. Sensors 2017, 17, 578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Ren, H.; Cao, Z.; Tian, Y.; Cao, W.; Zhu, Y.; Cheng, T. Detecting leaf nitrogen content in wheat with canopy hyperspectral under different soil backgrounds. Int. J. Appl. Earth Obs. Geoinf. 2014, 32, 114–124. [Google Scholar] [CrossRef]
- Hansen, P.; Schjoerring, J.K. Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote. Sens. Environ. 2003, 86, 542–553. [Google Scholar] [CrossRef]
- Chen, P.; Haboudane, D.; Tremblay, N.; Wang, J.; Vigneault, P.; Li, B. New spectral indicator assessing the efficiency of crop nitrogen treatment in corn and wheat. Remote. Sens. Environ. 2010, 114, 1987–1997. [Google Scholar] [CrossRef]
- Adams, M.L.; Philpot, W.D.; Norvell, W.A. Yellowness index: An application of spectral second derivatives to estimate chlorosis of leaves in stressed vegetation. Int. J. Remote. Sens. 1999, 20, 3663–3675. [Google Scholar] [CrossRef]
- Serrano, L.; Penuelas, J.; Ustin, S. Remote sensing of nitrogen and lignin in Mediterranean vegetation from AVIRIS data: De-composing biochemical from structural signals. Remote Sens. Environ. 2002, 81, 355–364. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote. Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Huete, A. A soil-adjusted vegetation index (SAVI). Remote. Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Fitzgerald, G.; Rodriguez, D.; Christensen, L.K.; Belford, R.; Sadras, V.O.; Clarke, T.R. Spectral and thermal sensing for nitrogen and water status in rainfed and irrigated wheat environments. Precis. Agric. 2006, 7, 233–248. [Google Scholar] [CrossRef]
- Richardson, A.; Wiegand, C. Distinguishing vegetation from soil background information. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]
- Liu, H.; Huete, A. A feedback based modification of the NDVI to minimize canopy background and atmospheric noise. IEEE Trans. Geosci. Remote Sens. 1995, 33, 457–465. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr.; Haas, R.; Schell, J.; Deering, D. Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote. Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Schuerger, A.C.; Capelle, G.A.; Di Benedetto, J.A.; Mao, C.; Thai, C.N.; Evans, M.D.; Richards, J.T.; A Blank, T.; Stryjewski, E.C. Comparison of two hyperspectral imaging and two laser-induced fluorescence instruments for the detection of zinc stress and chlorophyll concentration in bahia grass (Paspalum notatum Flugge.). Remote. Sens. Environ. 2003, 84, 572–588. [Google Scholar] [CrossRef]
- Tang, S.; Zhu, Q.; Wang, J.; Zhou, Y.; Zhao, F. Theoretical bases and application of three gradient difference vegetation index. Sci. China Ser. D 2003, 33, 1094–1102. [Google Scholar]
- Broge, N.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for esti-mation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.; Pattey, E.; Zarco-Tejada, P.; Strachan, I. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote. Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of Vegetation Indices and a Modified Simple Ratio for Boreal Applications. Can. J. Remote. Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Kaufman, Y.J.; Tanre, D. Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. IEEE Trans. Geosci. Remote. Sens. 1992, 30, 261–270. [Google Scholar] [CrossRef]
- Vogelmann, J.E.; Rock, B.N.; Moss, D.M. Red edge spectral measurements from sugar maple leaves. Int. J. Remote Sens. 1993, 14, 1563–1575. [Google Scholar] [CrossRef]
- Gamon, J.; Peñuelas, J.; Field, C. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote. Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Gamon, J.A.; Serrano, L.; Surfus, J.S. The photochemical reflectance index: An optical indicator of photosynthetic radiation use efficiency across species, functional types, and nutrient levels. Oecologia 1997, 112, 492–501. [Google Scholar] [CrossRef]
- Peñuelas, J.; Gamon, J.; Fredeen, A.; Merino, J.; Field, C. Reflectance indices associated with physiological changes in nitro-gen-and water-limited sunflower leaves. Remote Sens. Environ. 1994, 48, 135–146. [Google Scholar] [CrossRef]
- Penuelas, J.; Baret, F.; Filella, I. Semi-empirical indices to assess carotenoids/chlorophyll a ratio from leaf spectral reflectance. Photosynthetica 1995, 31, 221–230. [Google Scholar]
- Merzlyak, M.N.; Gitelson, A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef] [Green Version]
- Strachan, I.; Pattey, E.; Boisvert, J. Impact of nitrogen and environmental conditions on corn as detected by hyperspectral re-flectance. Remote Sens. Environ. 2002, 80, 213–224. [Google Scholar] [CrossRef]
- Fan, L.; Zhao, J.; Xu, X.; Liang, D.; Yang, G.; Feng, H.; Yang, H.; Wang, Y.; Chen, G.; Wei, P. Hyperspectral-based Estimation of Leaf Nitrogen Content in Corn Using Optimal Selection of Multiple Spectral Variables. Sensors 2019, 19, 2898. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Yang, G.; Wang, J.; Song, X.; Feng, H. Winter wheat biomass estimation based on spectral indices, band depth analysis and partial least squares regression using hyperspectral measurements. Comput. Electron. Agric. 2014, 100, 51–59. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM. 2017, 6, 84–90. [Google Scholar] [CrossRef]
- Rigatti, S.J. Random Forest. J. Insur. Med. 2017, 47, 31–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef] [Green Version]
- Höskuldsson, A. PLS regression methods. J. Chemom. 1988, 2, 211–228. [Google Scholar] [CrossRef]
- Lu, C.-J.; Lee, T.-S.; Chiu, C.-C. Financial time series forecasting using independent component analysis and support vector regression. Decis. Support Syst. 2009, 47, 115–125. [Google Scholar] [CrossRef]
- Joharestani, M.Z.; Cao, C.; Ni, X.; Bashir, B.; Talebiesfandarani, S. PM2.5 Prediction Based on Random Forest, XGBoost, and Deep Learning Using Multisource Remote Sensing Data. Atmosphere 2019, 10, 373. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Qi, L.; Wang, M.; Hussain, S.; Wang, H.; Wang, B.; Ning, J. Cross-Category Tea Polyphenols Evaluation Model Based on Feature Fusion of Electronic Nose and Hyperspectral Imagery. Sensors 2020, 20, 50. [Google Scholar] [CrossRef] [Green Version]
- Davide, C.; Glenn, F.; Raffaele, C.; Bruno, B. Assessing the robustness of vegetation indices to estimate wheat N in Mediterranean environments. Remote Sens. 2014, 6, 2827–2844. [Google Scholar]
- Iglesias-Puzas, Á.; Boixeda, P. Deep Learning and Mathematical Models in Dermatology. Actas Dermo-Sifiliogr. 2020, 111, 192–195. [Google Scholar] [CrossRef]
- Cheng, G.; Li, Z.; Han, J.W.; Yao, X.W.; Guo, L. Exploring hierarchical convolutional features for hyperspectral image classifica-tion. IEEE Trans. Geosci. Remote Sens. 2018, 56, 6712–6722. [Google Scholar] [CrossRef]
- Mirzaei, A.; Pourahmadi, V.; Soltani, M.; Sheikhzadeh, H. Deep feature selection using a teacher-student net-work. Neurocomputing 2020, 383, 396–408. [Google Scholar] [CrossRef] [Green Version]
- Jia, M.; Li, W.; Wang, K.; Zhou, C.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; Yao, X. A newly developed method to extract the optimal hyperspectral feature for monitoring leaf biomass in wheat. Comput. Electron. Agric. 2019, 165, 104942. [Google Scholar] [CrossRef]







| Index | Formula | Reference |
|---|---|---|
| [35] | ||
| [36] | ||
| [37] | ||
| [38] | ||
| [39] | ||
| [40] | ||
| [41] | ||
| [42] | ||
| [43] | ||
| [44] | ||
| [45] | ||
| [46] | ||
| [47] | ||
| [48] | ||
| [49] | ||
| [50] | ||
| [51] | ||
| [52] | ||
| [53] | ||
| [54] | ||
| [54] | ||
| [54] | ||
| [55,56] | ||
| [57] | ||
| [58] | ||
| [59] |
| Variables | Calculation Formula |
|---|---|
| Variables | Names | Definition and Description |
|---|---|---|
| Blue edge amplitude | Maximum value of the 1st derivative of a blue edge (490–530 nm) | |
| Blue edge position | Wavelength at Db | |
| Yellow edge amplitude | Maximum value of the 1st derivative of a yellow edge (560–640 nm) | |
| Yellow edge position | Wavelength at Dy | |
| Red edge amplitude | Maximum value of the 1st derivative with a red edge (680–760 nm) | |
| Red edge position | Wavelength at Dr | |
| Green peak amplitude | Maximum reflectance of a green peak (510–560 nm) | |
| Location of green peak | Wavelength at Rg | |
| Red valley amplitude | Lowest reflectance of a red well (650–690 nm) | |
| Red valley position | Wavelength at Ro | |
| Blue-edge integral areas | Sum of the 1st derivative values within the blue edge | |
| Yellow-edge integral areas | Sum of the 1st derivative values within the yellow edge | |
| Red-edge integral areas | Sum of the 1st derivative values within the red well |
| Model | Features | Preferred | Calibration Set | Validation Set | ||
|---|---|---|---|---|---|---|
| Variables | R2 | RMSE | R2 | RMSE | ||
| PLS | VIs | 8 | 0.791 | 0.448 | 0.708 | 0.439 |
| PFs | 7 | 0.812 | 0.421 | 0.722 | 0.392 | |
| DFs | 20 | 0.867 | 0.352 | 0.794 | 0.330 | |
| FFs | 35 | 0.895 | 0.313 | 0.814 | 0.328 | |
| SVR | VIs | 8 | 0.791 | 0.442 | 0.659 | 0.449 |
| PFs | 7 | 0.809 | 0.448 | 0.703 | 0.416 | |
| DFs | 20 | 0.897 | 0.325 | 0.780 | 0.367 | |
| FFs | 35 | 0.954 | 0.209 | 0.842 | 0.312 | |
| GBDT | VIs | 8 | 0.848 | 0.148 | 0.717 | 0.384 |
| PFs | 7 | 0.853 | 0.137 | 0.77 | 0.386 | |
| DFs | 20 | 0.927 | 0.084 | 0.832 | 0.303 | |
| FFs | 35 | 0.975 | 0.01 | 0.861 | 0.263 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, B.; Ma, J.; Yao, X.; Cao, W.; Zhu, Y. Estimation of Leaf Nitrogen Content in Wheat Based on Fusion of Spectral Features and Deep Features from Near Infrared Hyperspectral Imagery. Sensors 2021, 21, 613. https://0-doi-org.brum.beds.ac.uk/10.3390/s21020613
Yang B, Ma J, Yao X, Cao W, Zhu Y. Estimation of Leaf Nitrogen Content in Wheat Based on Fusion of Spectral Features and Deep Features from Near Infrared Hyperspectral Imagery. Sensors. 2021; 21(2):613. https://0-doi-org.brum.beds.ac.uk/10.3390/s21020613
Chicago/Turabian StyleYang, Baohua, Jifeng Ma, Xia Yao, Weixing Cao, and Yan Zhu. 2021. "Estimation of Leaf Nitrogen Content in Wheat Based on Fusion of Spectral Features and Deep Features from Near Infrared Hyperspectral Imagery" Sensors 21, no. 2: 613. https://0-doi-org.brum.beds.ac.uk/10.3390/s21020613