Folate-Targeted mRNA Delivery Using Chitosan-Functionalized Selenium Nanoparticles: Potential in Cancer Immunotherapy
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation and Characterization of Nanoparticles
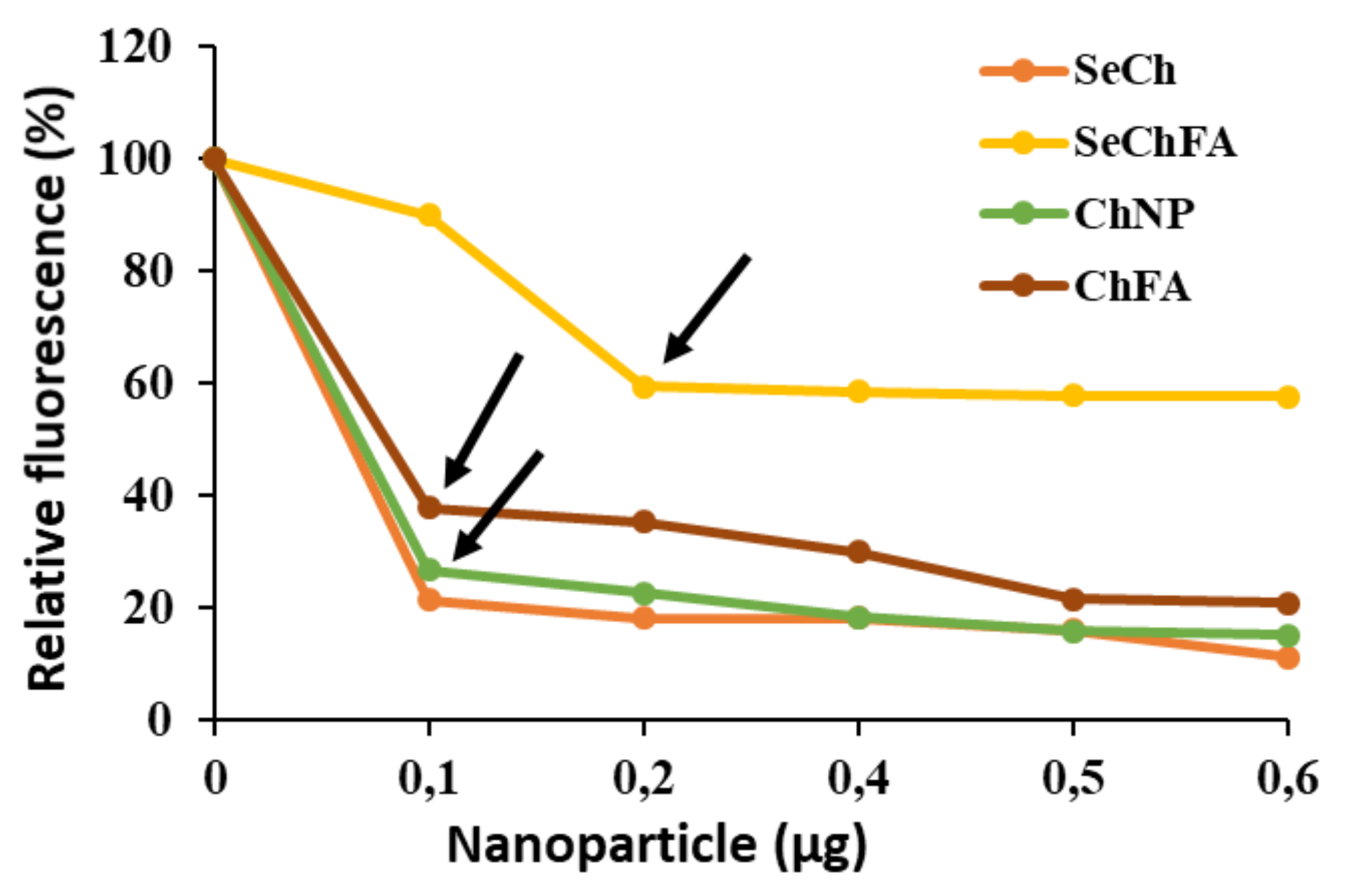
2.2. mRNA Binding and Nuclease Protection Studies
2.3. Cytotoxicity Studies
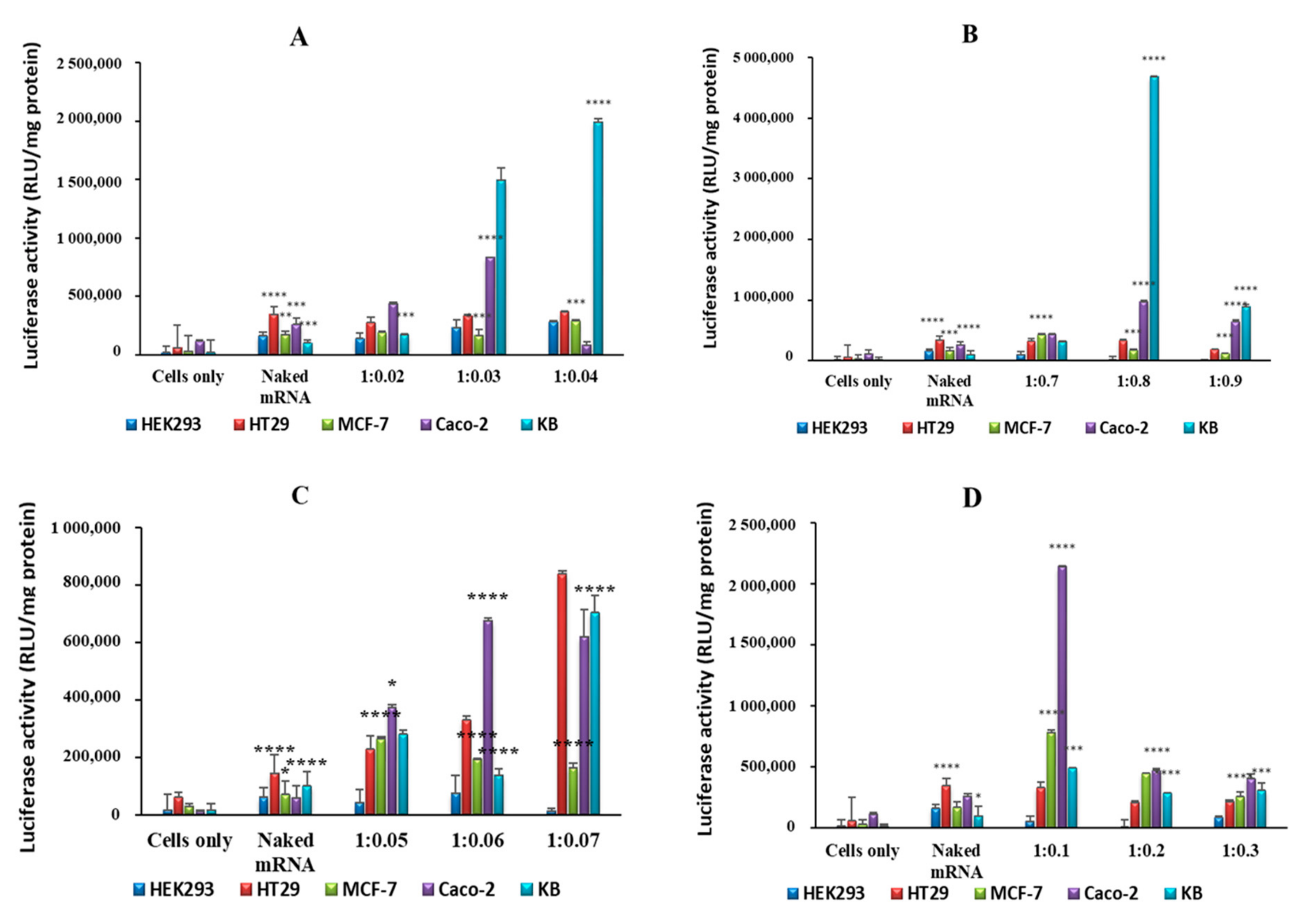
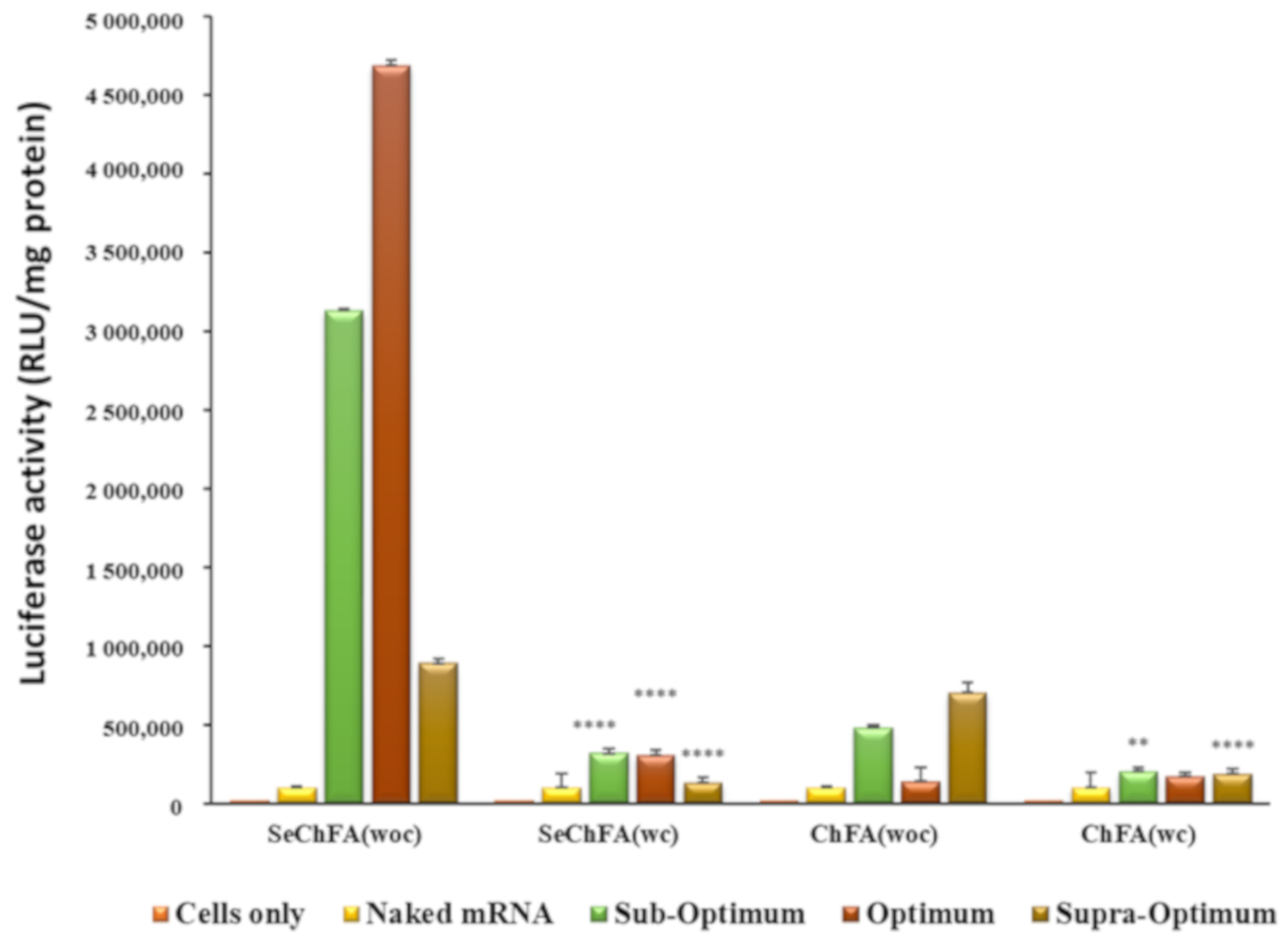
2.4. In Vitro Luciferase Expression
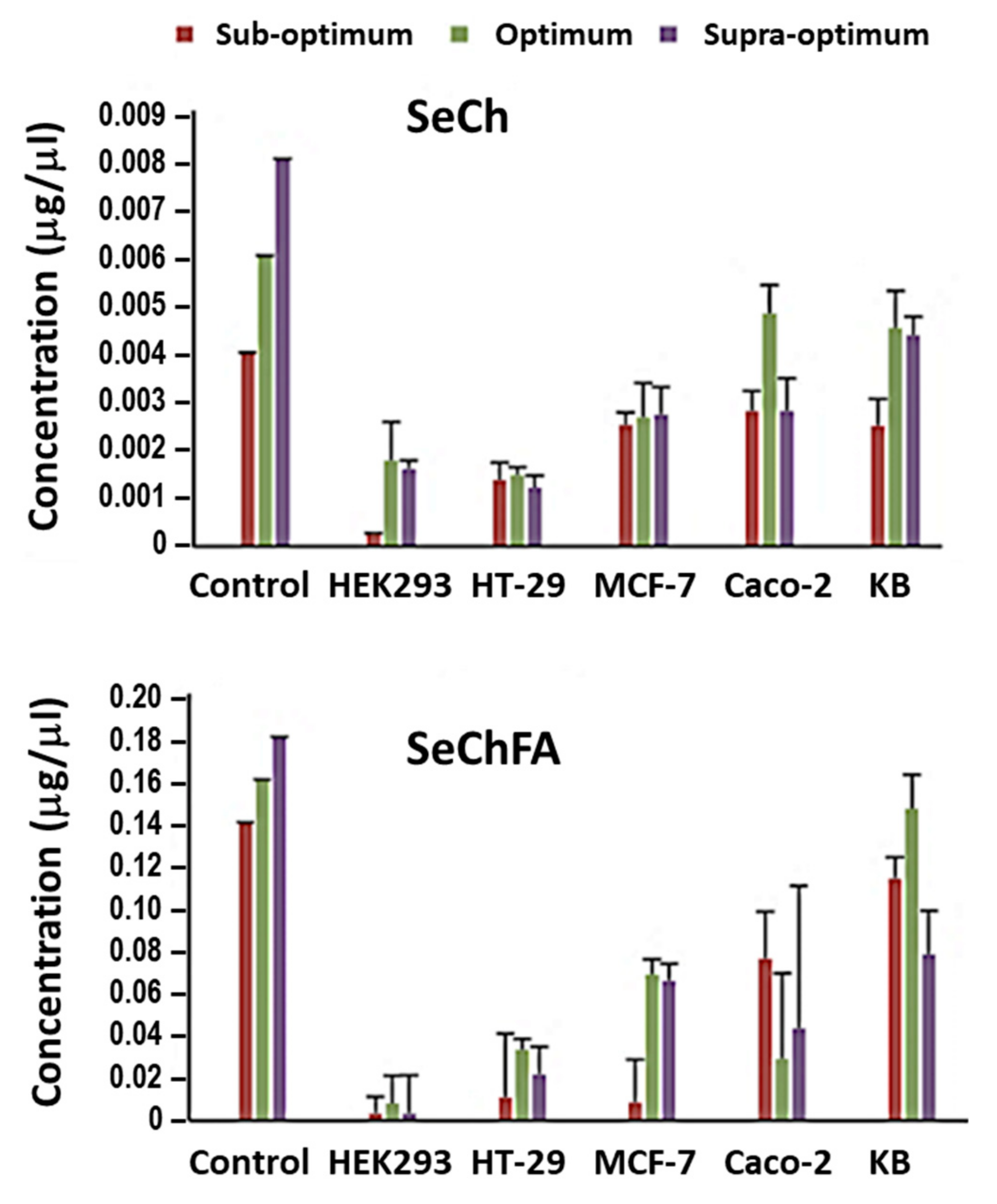
2.5. Selenium Uptake
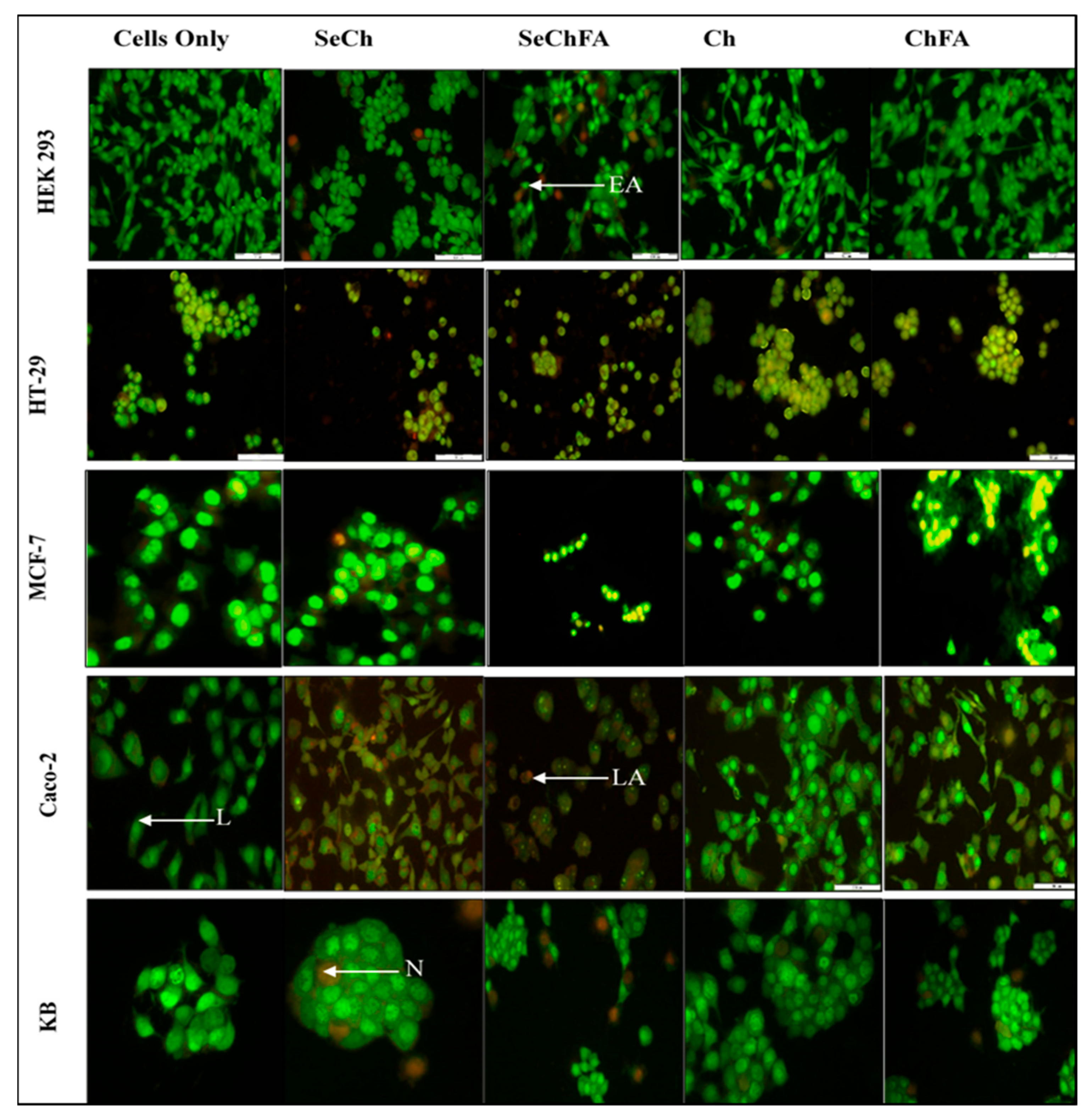
2.6. AO/EB Dual Staining
3. Materials and Methods
3.1. Materials
3.2. Preparation and Modification of Selenium Nanoparticles (SeNPs)
3.3. mRNA/NP Binding
3.4. Ethidium Bromide Intercalation Assay
3.5. RNase A Protection Assay
3.6. Nanoparticle and Nanocomplex Characterization
3.7. Cell Viability Assays
3.8. Gene Expression
3.9. Selenium Uptake
3.10. Acridine Orange/Ethidium Bromide Dual Staining
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amer, M.H. Gene therapy for cancer: Present status and future perspective. Mol. Cell. Ther. 2014, 2, 27. [Google Scholar] [CrossRef] [PubMed]
- Debus, H.; Baumhof, P.; Probst, J.; Kissel, T. Delivery of messenger RNA using poly (ethylene-imine)-poly (ethylene glycol)-copolymer blends for polyplex formation: Biophysical characterization and in vitro transfection properties. J. Control. Release 2010, 148, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Tolmachov, O.E.; Tolmachova, T. Methods of Transfection with Messenger RNA Gene Vectors. In Gene Therapy-Principles and Challenges; Hashad, D., Ed.; In TechOpen: London, UK, 2015; pp. 37–55. [Google Scholar]
- Tavernier, G.; Andries, O.; Demeester, J.; Sanders, N.N.; De Smedt, S.C.; Rejman, J. mRNA as gene therapeutic: How to control protein expression. J. Control. Release 2011, 150, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Moon, J.J. Nanoparticle drug delivery systems designed to improve cancer vaccines and immunotherapy. Vaccines 2015, 3, 662–685. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kormann, M.; Rosenecker, J.; Rudolph, C. Current prospects for mRNA gene delivery. Eur. J. Pharm. Biopharm. 2009, 71, 484–4899. [Google Scholar] [CrossRef]
- Uchida, S.; Kataoka, K.; Itaka, K. Screening of mRNA chemical modification to maximize protein expression with reduced immunogenicity. Pharmaceutics 2015, 7, 137–151. [Google Scholar] [CrossRef]
- Avci-Adali, M.; Behring, A.; Keller, T.; Krajewski, S.; Schlensak, C.; Wendel, H.P. Optimized conditions for successful transfection of human endothelial cells with in vitro synthesized and modified mRNA for induction of protein expression. J. Biol. Eng. 2014, 8, 8. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg Robert, A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Youn, H.; Chung, J.K. Modified mRNA as an alternative to plasmid DNA (pDNA) for transcript replacement and vaccination therapy. Expert Opin. Biol. Ther. 2015, 15, 1337–1348. [Google Scholar] [CrossRef]
- Mockey, M.; Gonçalves, C.; Dupuy, F.P.; Lemoine, F.M.; Pichon, C.; Midoux, P. mRNA transfection of dendritic cells: Synergistic effect of ARCA mRNA capping with Poly (A) chains in cis and in trans for a high protein expression level. Biochem. Biophys. Res. Commun. 2006, 340, 1062–1068. [Google Scholar] [CrossRef]
- Ulmer, J.B.; Geall, A.J. Recent innovations in mRNA vaccines. Curr. Opin. Immunol. 2016, 41, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Weide, B.; Carralot, J.P.; Reese, A.; Scheel, B.; Eigentler, T.K.; Hoerr, I.; Rammensee, H.G.; Garbe, C.; Pascolo, S. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 2008, 31, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, K.; Sarkar, S.; Rao, K.J.; Paria, S. Core/shell nanoparticles in biomedical applications. Adv. Colloid Interface Sci. 2014, 209, 8–39. [Google Scholar] [CrossRef] [PubMed]
- Weekley, C.M.; Harris, H.H. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem. Soc. Rev. 2013, 42, 8870–8894. [Google Scholar] [CrossRef] [PubMed]
- Maney, V.; Singh, M. An in vitro assessment of Chitosan/Bimetallic PtAu nanocomposites as delivery vehicles for Doxorubicin. Nanomedicine 2017, 12, 2625–2640. [Google Scholar] [CrossRef] [PubMed]
- Maney, V.; Singh, M. The synergism of Platinum-Gold bimetallic nanoconjugates enhance 5-Fluorouracil delivery in vitro. Pharmaceutics 2019, 11, 439. [Google Scholar] [CrossRef] [PubMed]
- Maiyo, F.; Singh, M. Selenium Nanoparticles: Potential in Cancer Gene and Drug Delivery. Nanomedicine 2017, 12, 1075–1089. [Google Scholar] [CrossRef]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 33–41. [Google Scholar] [CrossRef]
- Tapiero, H.; Townsend, D.; Tew, K. The antioxidant role of selenium and seleno-compounds. Biomed. Pharmacother. 2003, 57, 134–141. [Google Scholar] [CrossRef]
- Yazdi, M.H.; Mahdavi, M.; Faghfouri, E.; Faramarzi, M.A.; Sepehrizadeh, Z.; Hassan, Z.M.; Gholami, M.; Shahverdi, A.R. Th1 immune response induction by biogenic selenium nanoparticles in mice with breast cancer: Preliminary vaccine model. Iran. J. Biotechnol. 2015, 13, 1–9. [Google Scholar] [CrossRef]
- Arthur, J.R.; McKenzie, R.C.; Beckett, G.J. Selenium in the immune system. J. Nutr. 2003, 133, 1457S–1459S. [Google Scholar] [CrossRef] [PubMed]
- Huang., Z.; Rose, A.H.; Hoffmann, P.R. The role of selenium in inflammation and immunity: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2012, 16, 705–743. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Sun, W.; Kissel, T. Chitosan-based formulations for delivery of DNA and siRNA. Adv. Drug Deliv. Rev. 2010, 62, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.Z.; Cai, L.L.; Li, J.; Zhao, M.D.; Chen, F.Y.; Yuan, H.; Hu, F. Receptor-mediated gene delivery by folic acid-modified stearic acid-grafted chitosan micelles. Int. J. Nanomed. 2011, 6, 1559–1568. [Google Scholar] [CrossRef] [Green Version]
- Akinyelu, J.; Singh, M. Chitosan stabilized Gold-Folate-Poly (lactide-co-glycolide) Nanoplexes Facilitate Efficient Gene Delivery in Hepatic and Breast Cancer Cells. J. Nanosci. Nanotechnol. 2018, 18, 4478–4486. [Google Scholar] [CrossRef]
- Daniels, A.N.; Singh, M. Sterically stabilised siRNA: Gold nanocomplexes enhance c-MYC silencing in a breast cancer cell model. Nanomedicine 2019, 14, 1387–1401. [Google Scholar] [CrossRef]
- Huang, Y.; He, L.; Liu, W.; Fan, C.; Zheng, W.; Wong, Y.S.; Chen, T. Selective cellular uptake and induction of apoptosis of cancer-targeted selenium nanoparticles. Biomaterials 2013, 34, 7106–7116. [Google Scholar] [CrossRef]
- Estevez, H.; Garcia-Lidon, J.C.; Luque-Garcia, J.L.; Camara, C. Effects of chitosan-stabilized selenium nanoparticles on cell proliferation, apoptosis and cell cycle pattern in HepG2 cells: Comparison with other selenospecies. Colloid. Surf. B 2014, 122, 184–193. [Google Scholar] [CrossRef]
- Zhai, X.; Zhang, C.; Zhao, G.; Stoll, S.; Ren, F.; Leng, X. Antioxidant capacities of the selenium nanoparticles stabilized by chitosan. J. Nanobiotechnol. 2017, 15, 4. [Google Scholar] [CrossRef]
- Liu, W.; Li, X.; Wong, Y.S.; Zheng, W.; Zhang, Y.; Cao, W.; Chen, T. Selenium nanoparticles as a carrier of 5-fluorouracil to achieve anticancer synergism. ACS Nano 2012, 6, 578–591. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Zeng, F.; Chen, L.; Peng, Z.; Kong, L.X. Synthesis and characterization of folate conjugated chitosan and cellular uptake of its nanoparticles in HT-29 cells. Carbohydr. Res. 2011, 346, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Li, X.; Zheng, W.; Feng, Y.; Wong, Y.S.; Chen, T. pH-responsive cancer-targeted selenium nanoparticles: A transformable drug carrier with enhanced theranostic effects. J. Mater. Chem. B 2014, 2, 5409–5418. [Google Scholar] [CrossRef]
- Esfandiarpour-Boroujeni, S.; Bagheri-Khoulenjani, S.; Mirzadeh, H. Modeling and optimization of degree of folate grafted on chitosan and carboxymethyl-chitosan. Progr. Biomater. 2016, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Furlani, F.; Sacco, P.; Marsich, E.; Donati, I.; Paoletti, S. Highly monodisperse colloidal coacervates based on bioactive lactose-modified chitosan: From synthesis to characterization. Carbohydr. Polym. 2017, 174, 360–368. [Google Scholar] [CrossRef]
- Roger, E.; Kalscheuer, S.; Kirtane, A.; Guru, B.R.; Grill, A.E.; Whittum-Hudson, J.; Panyam, J. Folic acid functionalized nanoparticles for enhanced oral drug delivery. Mol. Pharm. 2012, 9, 2103–2110. [Google Scholar] [CrossRef]
- Honary, S.; Zahir, F. Effect of zeta potential on the properties of nano-drug delivery systems-a review (Part 2). Trop. J. Pharm. Res. 2013, 12, 265–273. [Google Scholar]
- Zhang, S.; Luo, Y.; Zeng, H.; Wang, Q.; Tian, F.; Song, J.; Cheng, W.H. Encapsulation of selenium in chitosan nanoparticles improves selenium availability and protects cells from selenium-induced DNA damage response. J. Nutr. Biochem. 2011, 22, 1137–1142. [Google Scholar] [CrossRef]
- Lopez-Heras, I.; Sanchez-Diaz, R.; Anunciação, D.S.; Madrid, Y.; Luque-Garcia, J.L.; Camara, C. Effect of Chitosan-stabilized Selenium nanoparticles on cell cycle arrest and invasiveness in hepatocarcinoma cells revealed by quantitative proteomics. J. Nanomed. Nanotechnol. 2014, 5, 226. [Google Scholar] [CrossRef]
- Liu, K.; Liu, P.C.; Liu, R.; Wu, X. Dual AO/EB staining to detect apoptosis in osteosarcoma cells compared with flow cytometry. Med. Sci. Monit. Basic Res. 2015, 21, 15–20. [Google Scholar]
- Maiyo, F.; Moodley, R.; Singh, M. Cytotoxicity, antioxidant and apoptosis studies of quercetin-3-O glucoside and 4-(β-D-glucopyranosyl-1→4-α-L-rhamnopyranosyloxy)-benzyl isothiocyanate from Moringa oleifera. Anticancer Agents Med. Chem. 2016, 16, 648–656. [Google Scholar] [CrossRef]





| NP | Size (nm) | ζ Potential (mV) | PDI | Nanocomplexes at end-Point Ratios (Optimum Binding Ratio) | |||
|---|---|---|---|---|---|---|---|
| w/w | Size | ζ Potential | PDI | ||||
| Se | 85.3 ± 8 | −14.8 ± −3.6 | 0.00450 | - | - | - | |
| SeCH | 59.6 ± 0.1 | 21.0 ± 0.2 | 0 | 1:0.03 | 66.9 ± 0.9 | 14.4 ± 1.7 | 0.0025 |
| SeChFA | 75.6 ± 1.4 | 9.0 ± 0.3 | 0.00475 | 1:0.8 | 102.7 ± 15.2 | 9.8 ± 0.3 | 0.0006 |
| CH | 124.4 ± 19 | 38.5 ± 2.7 | 0.00064 | 1:0.06 | 162.9 ± 5.9 | 40.7 ± 1.3 | 0.0012 |
| ChFA | 139.5 ± 3.5 | 21.9 ± 0.7 | 0.00370 | 1:0.2 | 136.1 ± 18.5 | 32.9 ± 1.5 | 0.0009 |
| Cell Lines | Apoptotic Index | ||||
|---|---|---|---|---|---|
| Control | SeCh | SeChFA | Chitosan | ChFA | |
| HEK293 | 0 | 0 | 0.11 | 0 | 0 |
| HT29 | 0 | 0.15 | 0.09 | 0.11 | 0.23 |
| MCF7 | 0 | 0.07 | 0.28 | 0 | 0 |
| Caco-2 | 0 | 0.71 | 1 | 0.02 | 0.83 |
| KB | 0 | 0.04 | 0 | 0 | 0.02 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maiyo, F.; Singh, M. Folate-Targeted mRNA Delivery Using Chitosan-Functionalized Selenium Nanoparticles: Potential in Cancer Immunotherapy. Pharmaceuticals 2019, 12, 164. https://0-doi-org.brum.beds.ac.uk/10.3390/ph12040164
Maiyo F, Singh M. Folate-Targeted mRNA Delivery Using Chitosan-Functionalized Selenium Nanoparticles: Potential in Cancer Immunotherapy. Pharmaceuticals. 2019; 12(4):164. https://0-doi-org.brum.beds.ac.uk/10.3390/ph12040164
Chicago/Turabian StyleMaiyo, Fiona, and Moganavelli Singh. 2019. "Folate-Targeted mRNA Delivery Using Chitosan-Functionalized Selenium Nanoparticles: Potential in Cancer Immunotherapy" Pharmaceuticals 12, no. 4: 164. https://0-doi-org.brum.beds.ac.uk/10.3390/ph12040164






