Equine Mesenchymal Stem/Stromal Cells Freeze-Dried Secretome (Lyosecretome) for the Treatment of Musculoskeletal Diseases: Production Process Validation and Batch Release Test for Clinical Use
Abstract
:1. Introduction
2. Results
2.1. Lyosecretome Production
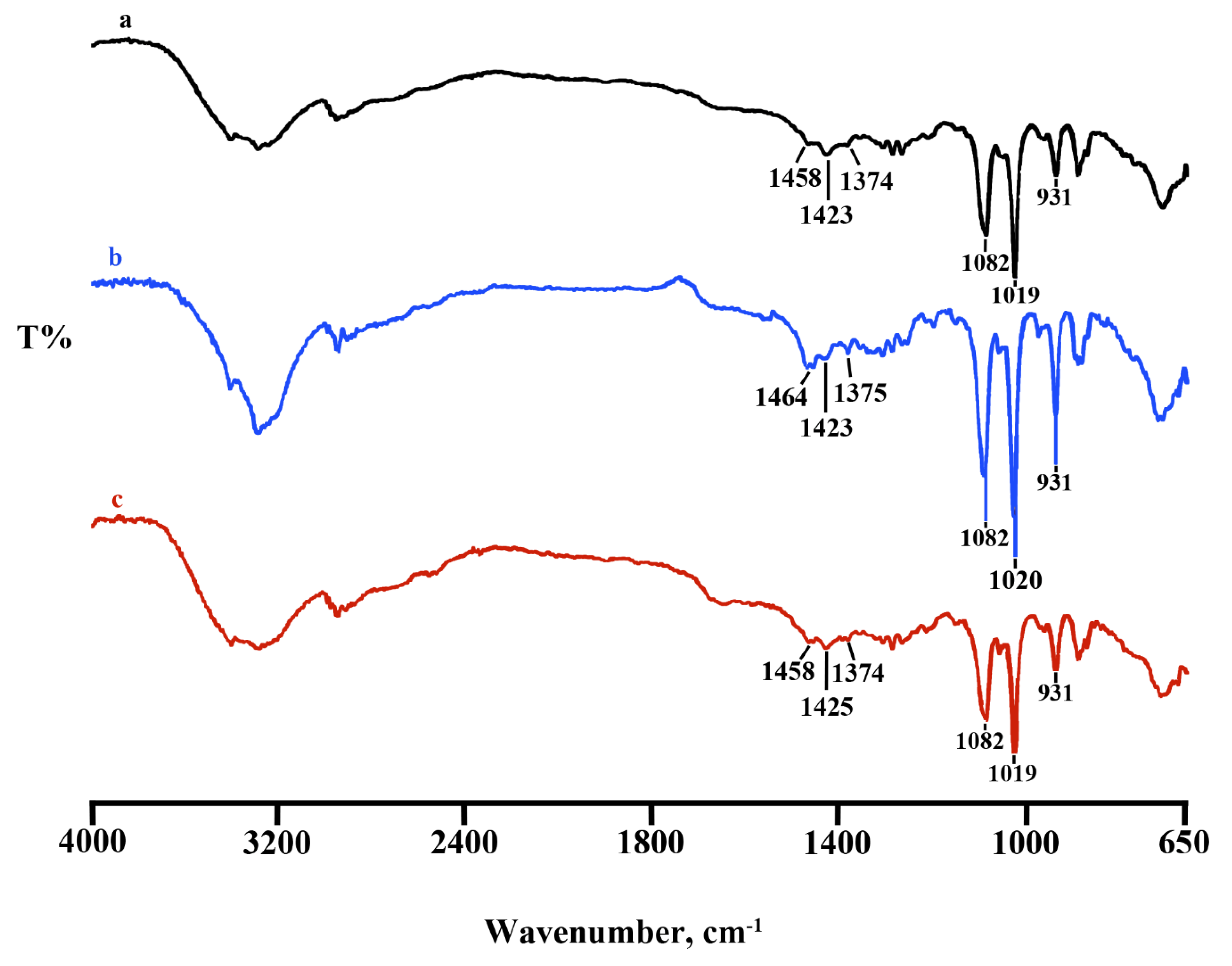
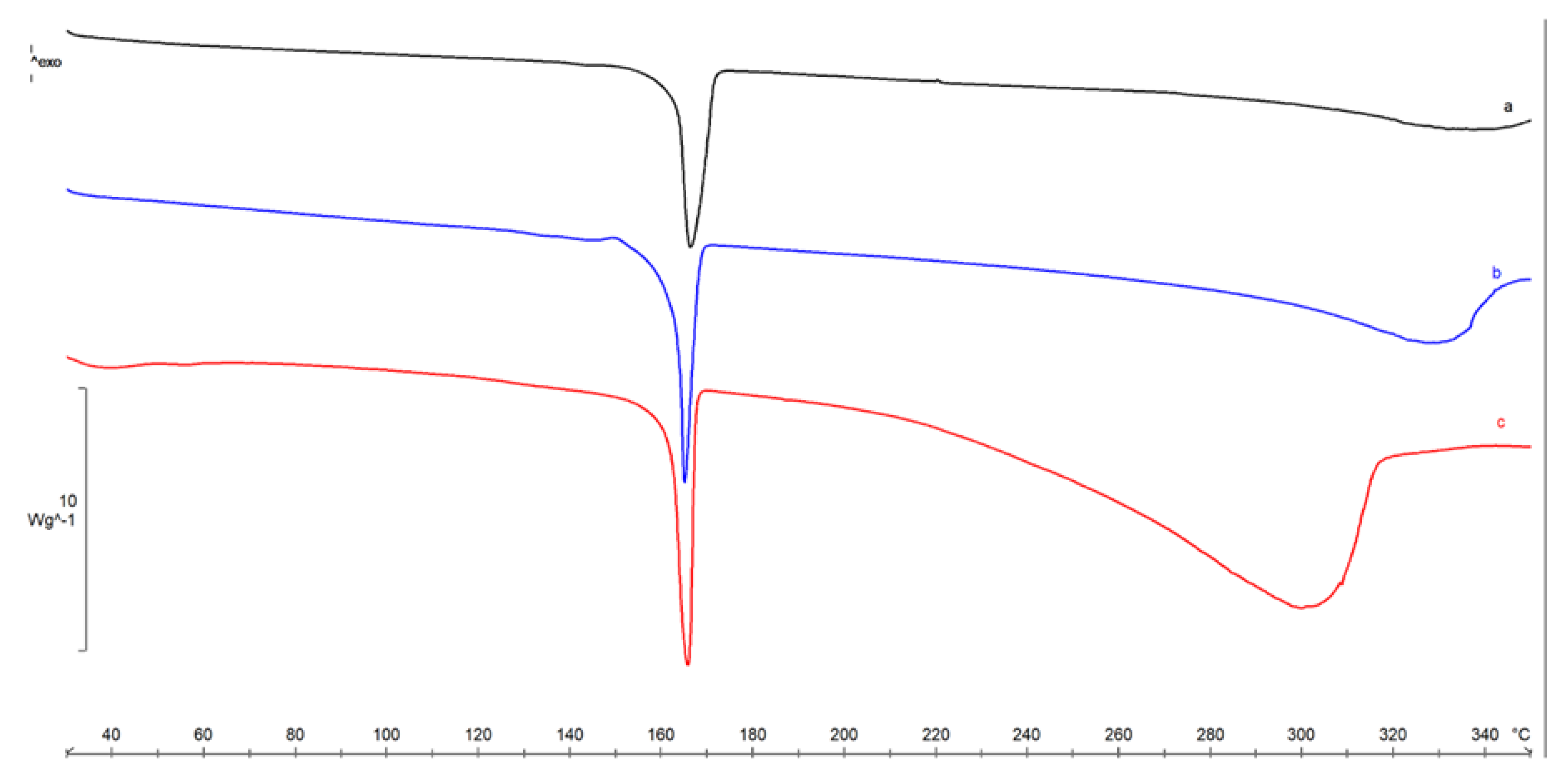
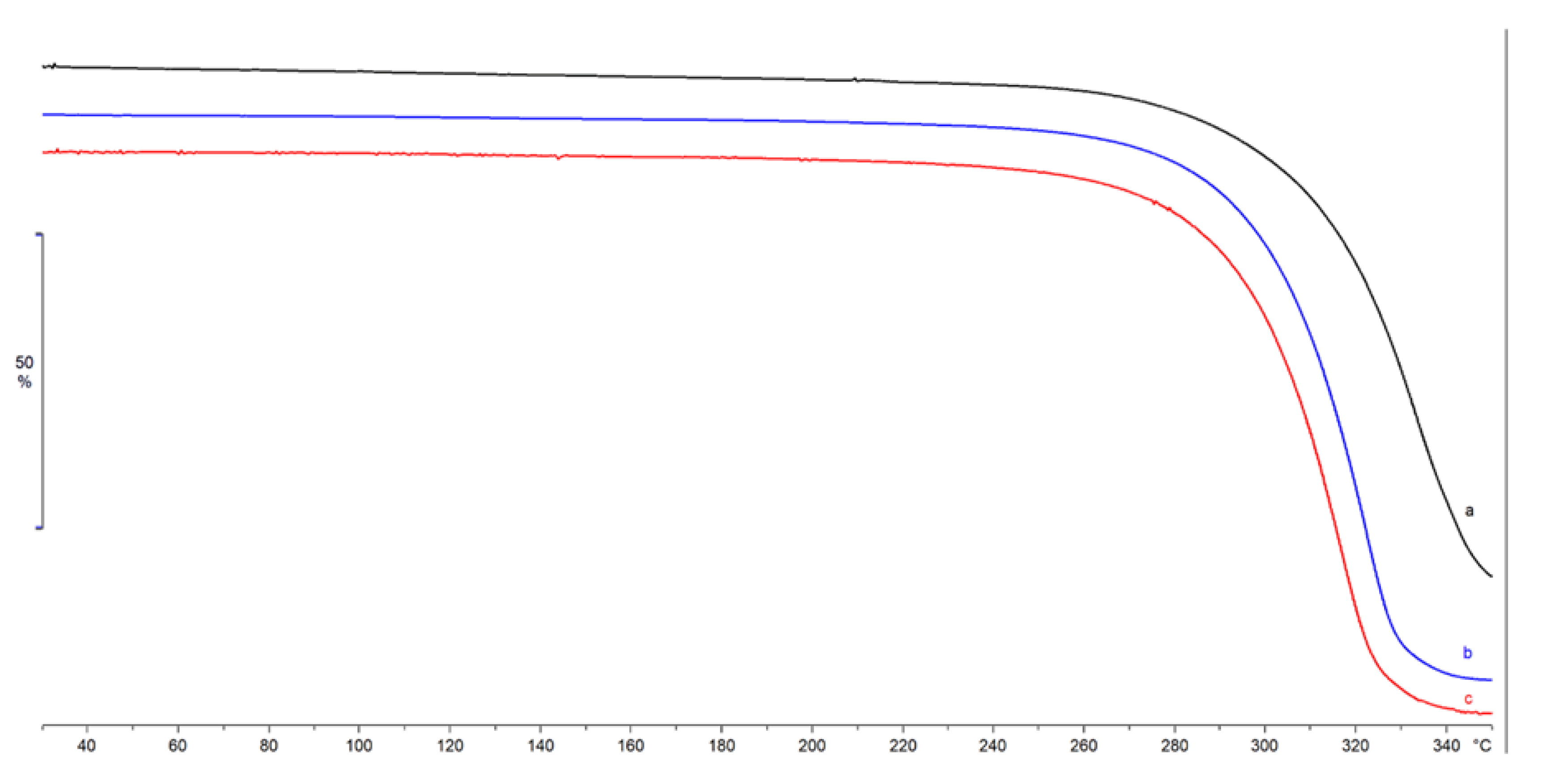
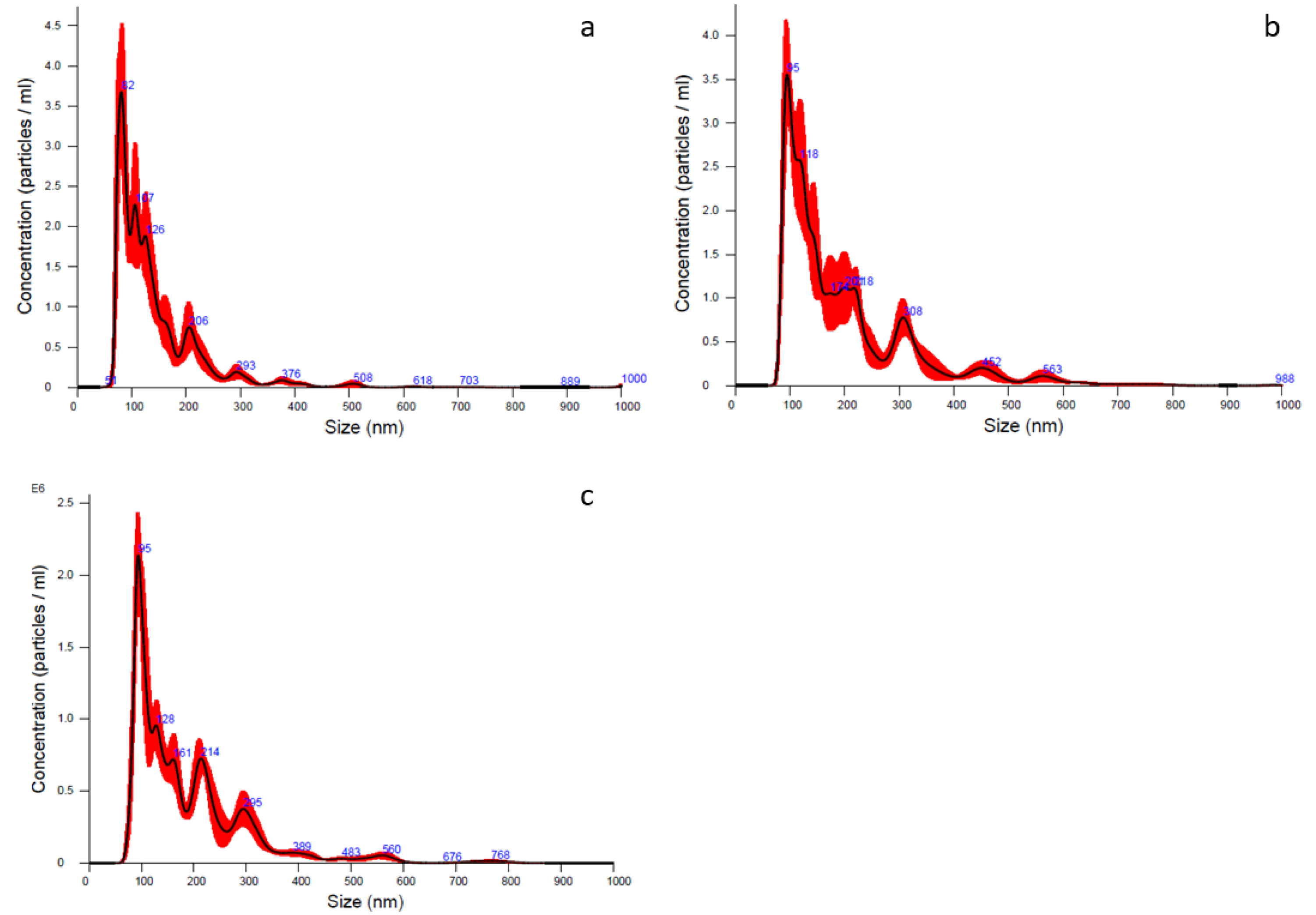
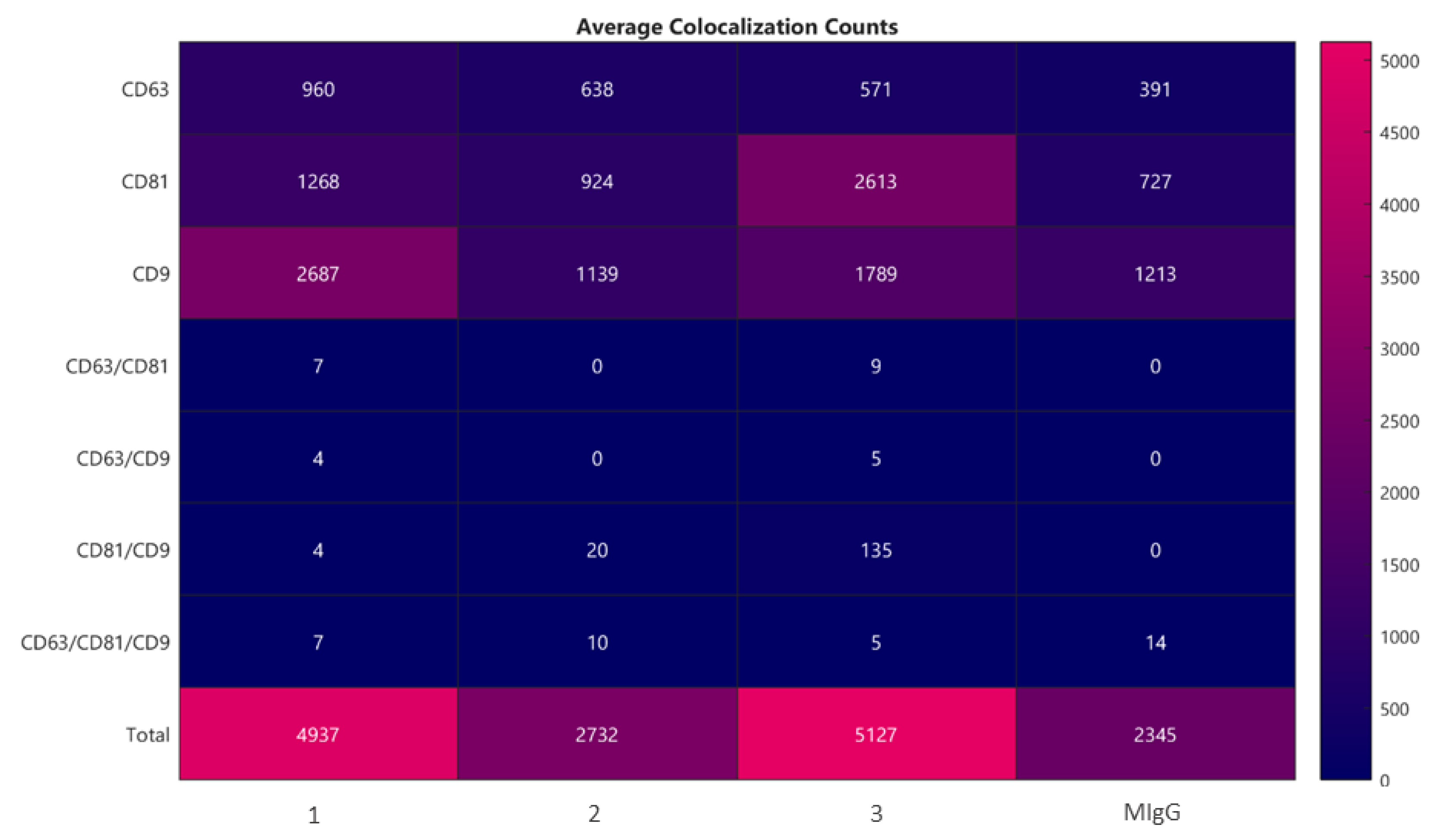
2.2. Lyosecretome Characterization
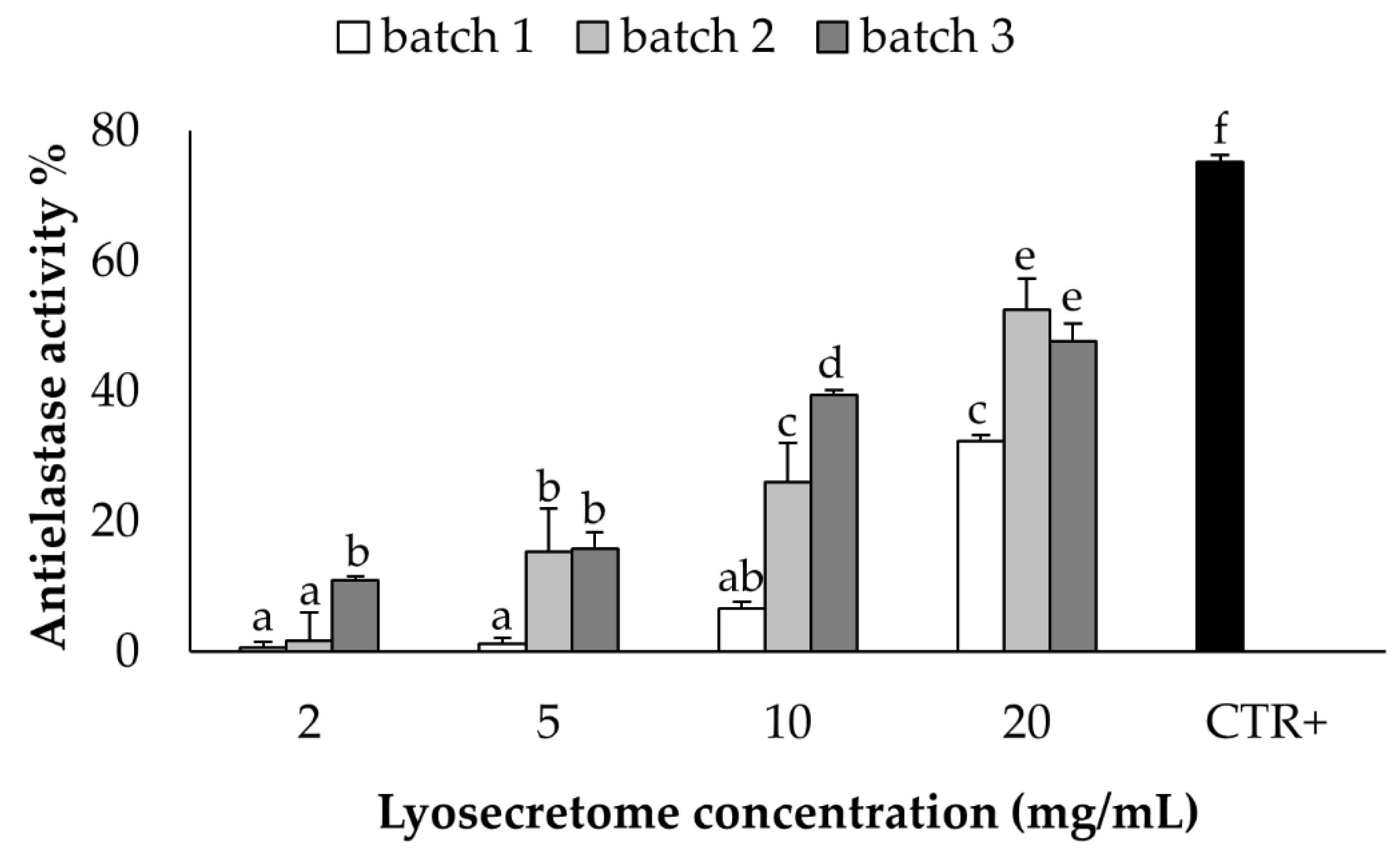
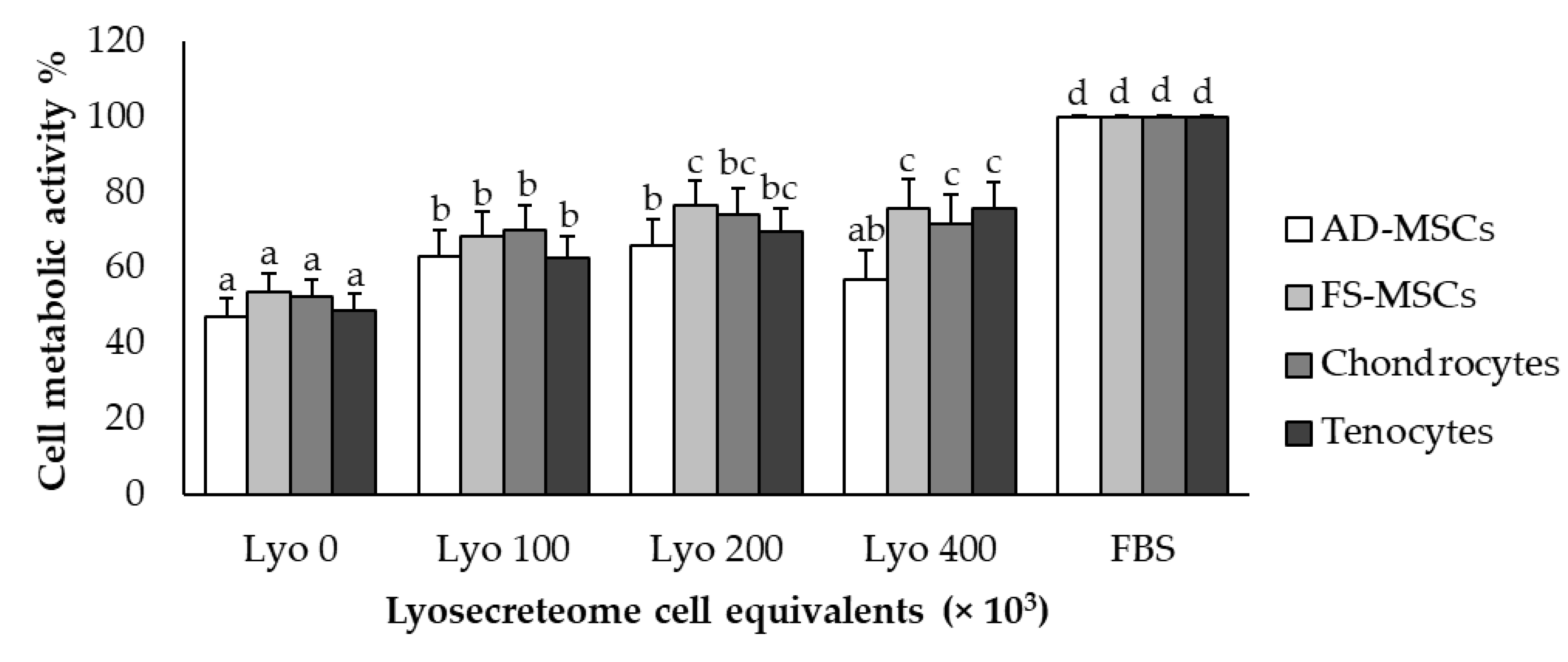
2.3. In Vitro Potency Test
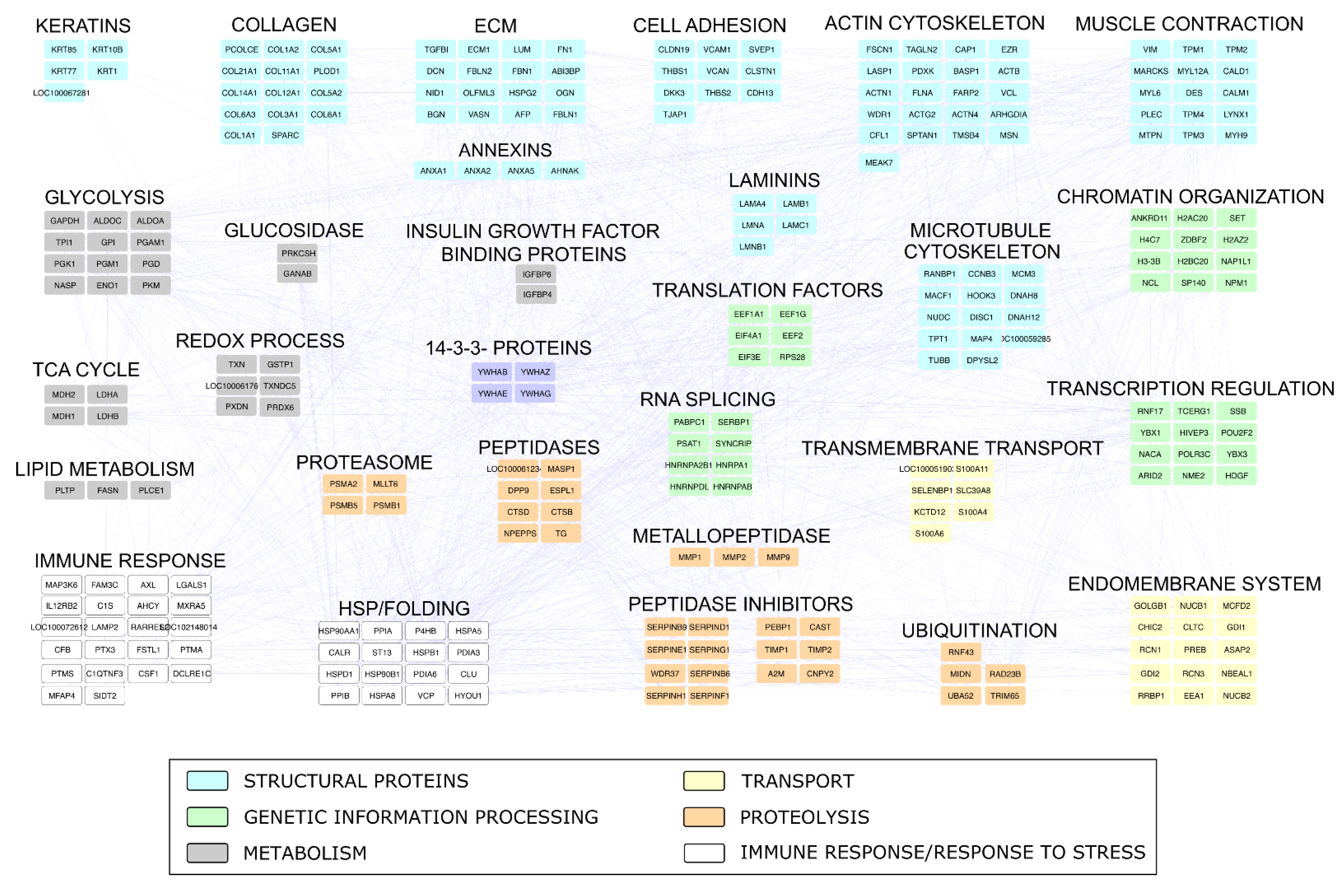
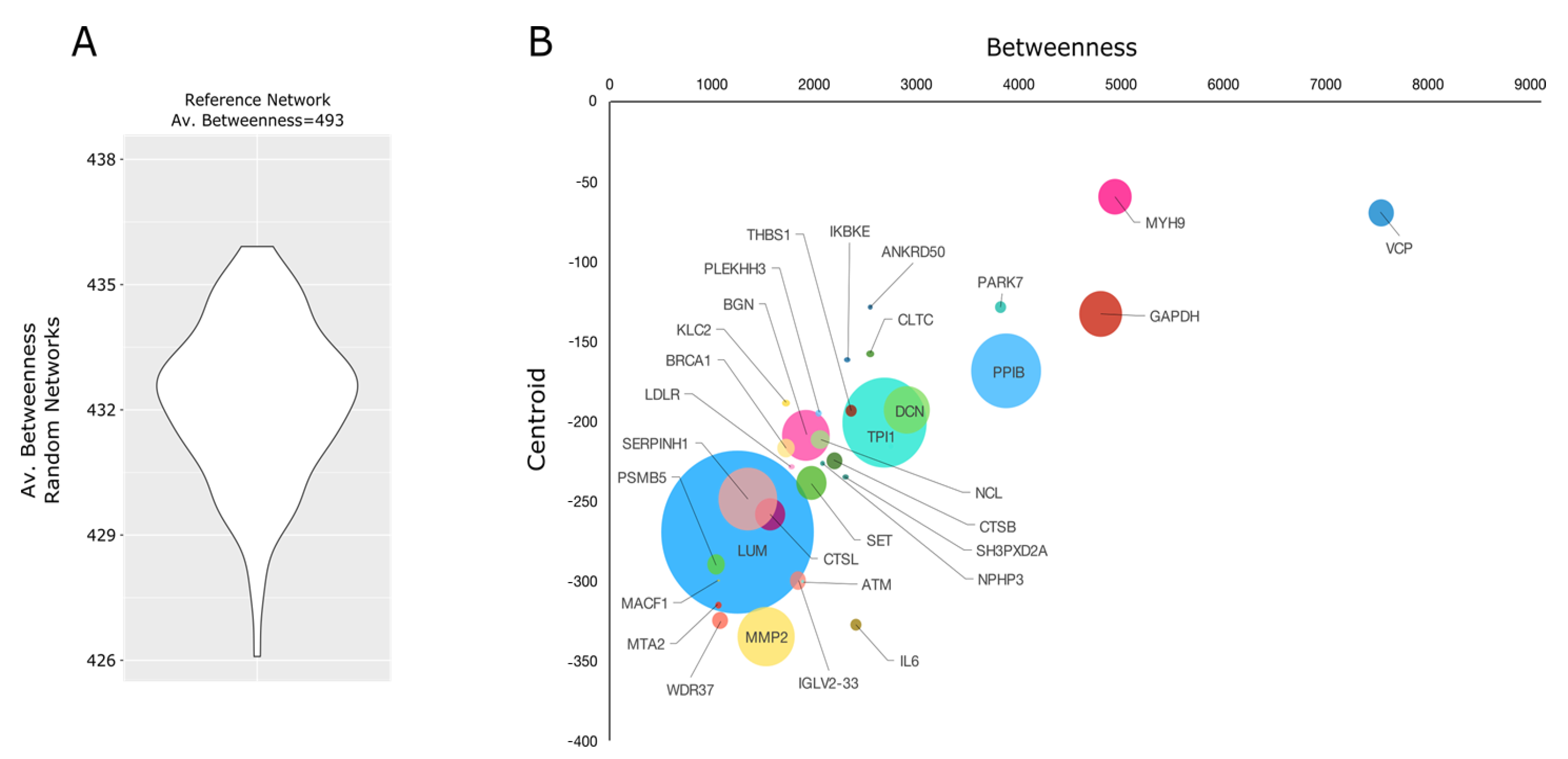
2.4. Proteomic Investigation
3. Discussion
4. Materials and Methods
4.1. Lyosecretome Production
4.1.1. Mesenchymal Stem/Stromal Cell Isolation
4.1.2. Mesenchymal Stem/Stromal Cell-Derived Secretome Production
4.1.3. Ultrafiltration
4.1.4. Secretome Freeze-Drying
4.2. Lyosecretome Characterization
4.2.1. Residual Humidity and Osmolarity
4.2.2. Fourier Transform Infrared Spectroscopy (FT-IR)
4.2.3. Thermal Characterization
4.2.4. Morphology Investigation by Scanning Electron Microscopy (SEM)
4.2.5. Particle Size Distribution
4.2.6. Microvesicles Surface Biomarkers by ExoView Analysis
4.2.7. Total Protein Quantification
4.2.8. Total Lipid Quantification
4.2.9. Sterility Test and Microbiological Control
4.3. In Vitro Potency Test
4.3.1. Elastase Inhibitory Assay
4.3.2. Proliferative Test
Synovial Fluid Cells Collection and Expansion
Tenocytes Isolation and Expansion
Chondrocytes Isolation and Expansion
Proliferative Test by MTT
4.4. Proteomic Investigation
4.4.1. LC-MS/MS Analysis
4.4.2. Proteomic Data Processing
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sevivas, N.; Teixeira, F.; Portugal, R.; Araújo, L.; Carriço, L.F.; Ferreira, N.V.; Da Silva, M.V.; Espregueira-Mendes, J.; Anjo, S.; Manadas, B.; et al. Mesenchymal Stem Cell Secretome: A Potential Tool for the Prevention of Muscle Degenerative Changes Associated with Chronic Rotator Cuff Tears. Am. J. Sports Med. 2016, 45, 179–188. [Google Scholar] [CrossRef]
- Galuzzi, M.; Perteghella, S.; Antonioli, B.; Tosca, M.C.; Bari, E.; Tripodo, G.; Sorrenti, M.; Catenacci, L.; Mastracci, L.; Grillo, F.; et al. Human Engineered Cartilage and Decellularized Matrix as an Alternative to Animal Osteoarthritis Model. Polymers 2018, 10, 738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martel-Pelletier, J.; Barr, A.J.; Cicuttini, F.M.; Conaghan, P.G.; Cooper, C.; Goldring, M.B.; Goldring, S.R.; Jones, G.; Teichtahl, A.J.; Pelletier, J.-P. Osteoarthritis. Nat. Rev. Dis. Prim. 2016, 2, 16072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McIlwraith, C.W.; Kawcak, C.; Baxter, G.M.; Goodrich, L.R.; Valberg, S.J. Joint injuries and disease and osteoarthritis. In Principles of Musculoskeletal Disease; Wiley: Hoboken, NJ, USA, 2020; Chapter 7. [Google Scholar] [CrossRef]
- Markoski, M.M. Advances in the Use of Stem Cells in Veterinary Medicine: From Basic Research to Clinical Practice. Science 2016, 2016, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, R.K.W.; Korda, M.; Blunn, G.W.; Goodship, A.E. Isolation and implantation of autologous equine mesenchymal stem cells from bone marrow into the superficial digital flexor tendon as a potential novel treatment. Equine Veter J. 2010, 35, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Lange-Consiglio, A.; Rossi, D.; Tassan, S.; Perego, R.; Cremonesi, F.; Parolini, O. Conditioned Medium from Horse Amniotic Membrane-Derived Multipotent Progenitor Cells: Immunomodulatory Activity In Vitro and First Clinical Application in Tendon and Ligament Injuries In Vivo. Stem Cells Dev. 2013, 22, 3015–3024. [Google Scholar] [CrossRef]
- Brandão, J.S.; Alvarenga, M.L.; Pfeifer, J.P.H.; Santos, V.H.; Fonseca-Alves, C.E.; Rodrigues, M.; Laufer-Amorim, R.; Castillo, J.A.L.; Alves, A.L.G.; De Souza, J.B. Allogeneic mesenchymal stem cell transplantation in healthy equine superficial digital flexor tendon: A study of the local inflammatory response. Res. Veter Sci. 2018, 118, 423–430. [Google Scholar] [CrossRef] [Green Version]
- Ricco, S.; Renzi, S.; Del Bue, M.; Conti, V.; Merli, E.; Ramoni, R.; Lucarelli, E.; Gnudi, G.; Ferrari, M.; Grolli, S. Allogeneic Adipose Tissue-Derived Mesenchymal Stem Cells in Combination with Platelet Rich Plasma are Safe and Effective in the Therapy of Superficial Digital Flexor Tendonitis in the Horse. Int. J. Immunopathol. Pharmacol. 2013, 26, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Abbasi-Malati, Z.; Roushandeh, A.M.; Kuwahara, Y.; Roudkenar, M.H. Mesenchymal Stem Cells on Horizon: A New Arsenal of Therapeutic Agents. Stem Cell Rev. Rep. 2018, 14, 484–499. [Google Scholar] [CrossRef]
- Konala, V.B.R.; Mamidi, M.K.; Bhonde, R.; Das, A.K.; Pochampally, R.; Pal, R. The current landscape of the mesenchymal stromal cell secretome: A new paradigm for cell-free regeneration. Cytotherapy 2016, 18, 13–24. [Google Scholar] [CrossRef] [Green Version]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Mocchi, M.; Dotti, S.; Del Bue, M.; Villa, R.; Bari, E.; Perteghella, S.; Torre, M.L.; Grolli, S. Veterinary Regenerative Medicine for Musculoskeletal Disorders: Can Mesenchymal Stem/Stromal Cells and Their Secretome Be the New Frontier? Cells 2020, 9, 1453. [Google Scholar] [CrossRef]
- Bari, E.; Perteghella, S.; Di Silvestre, D.; Sorlini, M.; Catenacci, L.; Sorrenti, M.; Marrubini, G.; Rossi, R.; Tripodo, G.; Mauri, P.; et al. Pilot Production of Mesenchymal Stem/Stromal Freeze-Dried Secretome for Cell-Free Regenerative Nanomedicine: A Validated GMP-Compliant Process. Cells 2018, 7, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orbay, H.; Tobita, M.; Mizuno, H. Mesenchymal Stem Cells Isolated from Adipose and Other Tissues: Basic Biological Properties and Clinical Applications. Stem Cells Int. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaetani, P.; Torre, M.L.; Klinger, M.; Faustini, M.; Crovato, F.; Bucco, M.; Marazzi, M.; Chlapanidas, T.; Levi, D.; Tancioni, F.; et al. Adipose-Derived Stem Cell Therapy for Intervertebral Disc Regeneration: AnIn VitroReconstructed Tissue in Alginate Capsules. Tissue Eng. Part A 2008, 14, 1415–1423. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sole, A.; Spriet, M.; Galuppo, L.D.; Padgett, K.A.; Borjesson, D.L.; Wisner, E.R.; Brosnan, R.J.; Vidal, M.A. Scintigraphic evaluation of intra-arterial and intravenous regional limb perfusion of allogeneic bone marrow-derived mesenchymal stem cells in the normal equine distal limb using 99mTc-HMPAO. Equine Veter J. 2012, 44, 594–599. [Google Scholar] [CrossRef]
- Smith, R.K.W.; Werling, N.; Dakin, S.; Alam, R.; Goodship, A.E.; Dudhia, J. Beneficial Effects of Autologous Bone Marrow-Derived Mesenchymal Stem Cells in Naturally Occurring Tendinopathy. PLoS ONE 2013, 8, e75697. [Google Scholar] [CrossRef]
- Franzè, S.; Selmin, F.; Samaritani, E.; Minghetti, P.; Cilurzo, F. Lyophilization of liposomal formulations: Still necessary, still challenging. Pharmaceutics 2018, 10, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, K.N.; Mallik, S.; Sarkar, K. Role of freeze-drying in the presence of mannitol on the echogenicity of echogenic liposomes. J. Acoust. Soc. Am. 2017, 142, 3670–3676. [Google Scholar] [CrossRef]
- Olsson, C.; Jansson, H.; Swenson, J. The Role of Trehalose for the Stabilization of Proteins. J. Phys. Chem. B 2016, 120, 4723–4731. [Google Scholar] [CrossRef] [PubMed]
- Demetzos, C. Differential Scanning Calorimetry (DSC): A Tool to Study the Thermal Behavior of Lipid Bilayers and Liposomal Stability. J. Liposome Res. 2008, 18, 159–173. [Google Scholar] [CrossRef]
- Han, S.; Li, Y.Y.; Chan, B.P. Protease inhibitors enhance extracellular collagen fibril deposition in human mesenchymal stem cells. Stem Cell Res. Ther. 2015, 6, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakrishnan, L.; Nirujogi, R.S.; Ahmad, S.; Bhattacharjee, M.; Manda, S.S.; Renuse, S.; Kelkar, D.S.; Subbannayya, Y.; Raju, R.; Goel, R.; et al. Proteomic analysis of human osteoarthritis synovial fluid. Clin. Proteom. 2014, 11, 6. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Chen, Q. Adipokines: New Therapeutic Target for Osteoarthritis? Curr. Rheumatol. Rep. 2019, 21, 71. [Google Scholar] [CrossRef] [PubMed]
- Berenbaum, F.; Eymard, F.; Houard, X. Osteoarthritis, inflammation and obesity. Curr. Opin. Rheumatol. 2013, 25, 114–118. [Google Scholar] [CrossRef]
- Pirozzi, C.; Francisco, V.; Di Guida, F.; Gómez, R.; Lago, F.; Pino, J.; Meli, R.; Gualillo, O. Butyrate Modulates Inflammation in Chondrocytes via GPR43 Receptor. Cell. Physiol. Biochem. 2018, 51, 228–243. [Google Scholar] [CrossRef]
- Poveda, J.; Sanz, A.B.; Fernandez-Fernandez, B.; Carrasco, S.; Ruiz-Ortega, M.; Cannata-Ortiz, P.; Ortiz, A.; Sanchez-Niño, M.D. MXRA5 is a TGF-β1-regulated human protein with anti-inflammatory and anti-fibrotic properties. J. Cell. Mol. Med. 2016, 21, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Ning, W.; Yu, Y.; Xu, H.; Liu, X.; Wang, D.; Wang, J.; Wang, Y.; Meng, W. The CAMSAP3-ACF7 Complex Couples Noncentrosomal Microtubules with Actin Filaments to Coordinate Their Dynamics. Dev. Cell 2016, 39, 61–74. [Google Scholar] [CrossRef] [Green Version]
- Zaoui, K.; Benseddik, K.; Daou, P.; Salaun, D.; Badache, A. ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc. Natl. Acad. Sci. USA 2010, 107, 18517–18522. [Google Scholar] [CrossRef] [Green Version]
- Saldaña, L.; Bensiamar, F.; Vallés, G.; Mancebo, F.J.; García-Rey, E.; Vilaboa, N. Immunoregulatory potential of mesenchymal stem cells following activation by macrophage-derived soluble factors. Stem Cell Res. Ther. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mohme, M.; Maire, C.L.; Geumann, U.; Schliffke, S.; Dührsen, L.; Fita, K.D.; Akyüz, N.; Binder, M.; Westphal, M.; Guenther, C.; et al. Local Intracerebral Immunomodulation Using Interleukin-Expressing Mesenchymal Stem Cells in Glioblastoma. Clin. Cancer Res. 2020, 26, 2626–2639. [Google Scholar] [CrossRef] [Green Version]
- Bundgaard, L.; Stensballe, A.; Elbæk, K.J.; Berg, L.C. Mass spectrometric analysis of the in vitro secretome from equine bone marrow-derived mesenchymal stromal cells to assess the effect of chondrogenic differentiation on response to interleukin-1β treatment. Stem Cell Res. Ther. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Borsini, A.; Di Benedetto, M.G.; Giacobbe, J.; Pariante, C.M. Pro- and Anti-Inflammatory Properties of Interleukin in Vitro: Relevance for Major Depression and Human Hippocampal Neurogenesis. Int. J. Neuropsychopharmacol. 2020, 23, 738–750. [Google Scholar] [CrossRef] [PubMed]
- Roseren, F.; Pithioux, M.; Robert, S.; Balasse, L.; Guillet, B.; Lamy, E.; Roffino, S. Systemic Administration of G-CSF Accelerates Bone Regeneration and Modulates Mobilization of Progenitor Cells in a Rat Model of Distraction Osteogenesis. Int. J. Mol. Sci. 2021, 22, 3505. [Google Scholar] [CrossRef] [PubMed]
- Otabe, K.; Nakahara, H.; Hasegawa, A.; Matsukawa, T.; Ayabe, F.; Onizuka, N.; Inui, M.; Takada, S.; Ito, Y.; Sekiya, I.; et al. Transcription factor Mohawk controls tenogenic differentiation of bone marrow mesenchymal stem cells in vitro and in vivo. J. Orthop. Res. 2015, 33, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Tornero-Esteban, P.; Peralta-Sastre, A.; Herranz, E.; Rodríguez-Rodríguez, L.; Mucientes, A.; Abásolo, L.; Marco, F.; Fernandez-Gutierrez, B.; Lamas, J.R. Altered Expression of Wnt Signaling Pathway Components in Osteogenesis of Mesenchymal Stem Cells in Osteoarthritis Patients. PLoS ONE 2015, 10, e0137170. [Google Scholar] [CrossRef]
- Bruno, S.; Grange, C.; Deregibus, M.C.; Calogero, R.; Saviozzi, S.; Collino, F.; Morando, L.; Busca, A.; Falda, M.; Bussolati, B.; et al. Mesenchymal Stem Cell-Derived Microvesicles Protect Against Acute Tubular Injury. J. Am. Soc. Nephrol. 2009, 20, 1053–1067. [Google Scholar] [CrossRef] [Green Version]
- Ni, G.-X.; Li, Z.; Zhou, Y.-Z. The role of small leucine-rich proteoglycans in osteoarthritis pathogenesis. Osteoarthr. Cartil. 2014, 22, 896–903. [Google Scholar] [CrossRef] [Green Version]
- Yazdanpanah, G.; Jiang, Y.; Rabiee, B.; Omidi, M.; Rosenblatt, M.I.; Shokuhfar, T.; Pan, Y.; Naba, A.; Djalilian, A.R. Fabrication, Rheological, and Compositional Characterization of Thermoresponsive Hydrogel from Cornea. Tissue Eng. Part C Methods 2021, 27, 307–321. [Google Scholar] [CrossRef]
- Karamanou, K.; Perrot, G.; Maquart, F.-X.; Brézillon, S. Lumican as a multivalent effector in wound healing. Adv. Drug Deliv. Rev. 2018, 129, 344–351. [Google Scholar] [CrossRef]
- Li, Q.; Han, B.; Wang, C.; Tong, W.; Wei, Y.; Tseng, W.; Han, L.; Liu, X.S.; Enomoto-Iwamoto, M.; Mauck, R.L.; et al. Mediation of Cartilage Matrix Degeneration and Fibrillation by Decorin in Post-traumatic Osteoarthritis. Arthritis Rheumatol. 2020, 72, 1266–1277. [Google Scholar] [CrossRef] [PubMed]
- Chery, D.R.; Han, B.; Zhou, Y.; Wang, C.; Adams, S.M.; Chandrasekaran, P.; Kwok, B.; Heo, S.-J.; Enomoto-Iwamoto, M.; Lu, X.L.; et al. Decorin regulates cartilage pericellular matrix micromechanobiology. Matrix Biol. 2021, 96, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Embree, M.C.; Kilts, T.M.; Ono, M.; Inkson, C.; Syed-Picard, F.; Karsdal, M.A.; Oldberg, Å.; Bi, Y.; Young, M.F. Biglycan and Fibromodulin Have Essential Roles in Regulating Chondrogenesis and Extracellular Matrix Turnover in Temporomandibular Joint Osteoarthritis. Am. J. Pathol. 2010, 176, 812–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bari, E.; Perteghella, S.; Catenacci, L.; Sorlini, M.; Croce, S.; Mantelli, M.; A Avanzini, M.; Sorrenti, M.; Torre, M.L. Freeze-dried and GMP-compliant pharmaceuticals containing exosomes for acellular mesenchymal stromal cell immunomodulant therapy. Nanomedicine 2019, 14, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Bari, E.; Ferrarotti, I.; Torre, M.L.; Corsico, A.G.; Perteghella, S. Mesenchymal stem/stromal cell secretome for lung regeneration: The long way through “pharmaceuticalization” for the best formulation. J. Control. Release 2019, 309, 11–24. [Google Scholar] [CrossRef]
- Mentkowski, K.; Snitzer, J.D.; Rusnak, S.; Lang, J.K. Therapeutic Potential of Engineered Extracellular Vesicles. AAPS J. 2018, 20, 50. [Google Scholar] [CrossRef] [Green Version]
- Orlandi, G.; Faragò, S.; Menato, S.; Sorlini, M.; Butti, F.; Mocchi, M.; Donelli, I.; Catenacci, L.; Sorrenti, M.L.; Croce, S.; et al. Eco-sustainable silk sericin from by-product of textile industry can be employed for cosmetic, dermatology and drug delivery. J. Chem. Technol. Biotechnol. 2020, 95, 2549–2560. [Google Scholar] [CrossRef]
- Orlandi, G.; Bari, E.; Catenacci, L.; Sorrenti, M.; Segale, L.; Faragò, S.; Sorlini, M.; Arciola, C.R.; Torre, M.L.; Perteghella, S. Polyphenols-Loaded Sericin Self-Assembling Nanoparticles: A Slow-Release for Regeneration by Tissue-Resident Mesenchymal Stem/Stromal Cells. Pharmaceutics 2020, 12, 381. [Google Scholar] [CrossRef] [Green Version]
- Washburn, M.P. The H-Index of ‘An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database’. J. Am. Soc. Mass Spectrom. 2015, 26, 1799–1803. [Google Scholar] [CrossRef] [Green Version]
- Deutsch, E.W.; Overall, C.M.; Van Eyk, J.E.; Baker, M.; Paik, Y.-K.; Weintraub, S.T.; Lane, L.; Martens, L.; Vandenbrouck, Y.; Kusebauch, U.; et al. Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 2.1. J. Proteome Res. 2016, 15, 3961–3970. [Google Scholar] [CrossRef] [PubMed]
- Motta, S.; Vecchietti, D.; Martorana, A.M.; Brunetti, P.; Bertoni, G.; Polissi, A.; Mauri, P.; Di Silvestre, D. The Landscape of Pseudomonas aeruginosa Membrane-Associated Proteins. Cells 2020, 9, 2421. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Di Silvestre, D.; Vigani, G.; Mauri, P.; Hammadi, S.; Morandini, P.; Murgia, I. Network Topological Analysis for the Identification of Novel Hubs in Plant Nutrition. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8.13.1–8.13.24. [Google Scholar] [CrossRef] [Green Version]
- Scardoni, G.; Tosadori, G.; Faizan, M.; Spoto, F.; Fabbri, F.; Laudanna, C. Biological network analysis with CentiScaPe: Centralities and experimental dataset integration. F1000Research 2014, 3, 139. [Google Scholar] [CrossRef] [Green Version]
- Sereni, L.; Castiello, M.C.; Di Silvestre, D.; Della Valle, P.; Brombin, C.; Ferrua, F.; Cicalese, M.P.; Pozzi, L.; Migliavacca, M.; Bernardo, M.E.; et al. Lentiviral gene therapy corrects platelet phenotype and function in patients with Wiskott-Aldrich syndrome. J. Allergy Clin. Immunol. 2019, 144, 825–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Batch n. | Total Cell Number × 106 | Cell Viability (%) |
|---|---|---|
| 1 | 224 | 99 |
| 2 | 170 | 98 |
| 3 | 65 | 99 |
| Batch n. | Sterility | Bacterial Endotoxin | Mycoplasma |
|---|---|---|---|
| 1 | compliant | 3.4 Eu/mL | no presence |
| 2 | compliant | 3.3 Eu/mL | no presence |
| 3 | compliant | 3.4 Eu/mL | no presence |
| Batch n. | Mean (nm) | Mode (nm) | d10 (nm) | d50 (nm) | d90 (nm) | Concentration (Particle/mL) |
|---|---|---|---|---|---|---|
| 1 | 142.7 ± 4.9 | 81.0 ± 2.8 | 77.5 ± 2.1 | 117.5 ± 3.2 | 231.2 ± 9.5 | 2.46 × 108 ± 1.48 × 107 |
| 2 | 198.5 ± 6.4 | 114.8 ± 11.8 | 94.8 ± 1.6 | 156.7 ± 8.6 | 253.5 ± 18.5 | 3.47 × 108 ± 2.66 × 107 |
| 3 | 187.1 ± 3.9 | 94.9 ± 3.0 | 91.5 ± 2.2 | 151.2 ± 2.8 | 261.3 ± 7.4 | 1.77 × 108 ± 7.77 × 106 |
| Batch n. | μg Proteins/mg Lyosecretome | μg Lipids/mg Lyosecretome |
|---|---|---|
| 1 | 10.0 ± 0.07 a | 0.7 ± 0.05 a |
| 2 | 13.0 ± 0.20 b | 1.0 ± 0.04 b |
| 3 | 10.6 ± 0.02 c | 0.6 ± 0.07 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mocchi, M.; Grolli, S.; Dotti, S.; Di Silvestre, D.; Villa, R.; Berni, P.; Conti, V.; Passignani, G.; Brambilla, F.; Bue, M.D.; et al. Equine Mesenchymal Stem/Stromal Cells Freeze-Dried Secretome (Lyosecretome) for the Treatment of Musculoskeletal Diseases: Production Process Validation and Batch Release Test for Clinical Use. Pharmaceuticals 2021, 14, 553. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14060553
Mocchi M, Grolli S, Dotti S, Di Silvestre D, Villa R, Berni P, Conti V, Passignani G, Brambilla F, Bue MD, et al. Equine Mesenchymal Stem/Stromal Cells Freeze-Dried Secretome (Lyosecretome) for the Treatment of Musculoskeletal Diseases: Production Process Validation and Batch Release Test for Clinical Use. Pharmaceuticals. 2021; 14(6):553. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14060553
Chicago/Turabian StyleMocchi, Michela, Stefano Grolli, Silvia Dotti, Dario Di Silvestre, Riccardo Villa, Priscilla Berni, Virna Conti, Giulia Passignani, Francesca Brambilla, Maurizio Del Bue, and et al. 2021. "Equine Mesenchymal Stem/Stromal Cells Freeze-Dried Secretome (Lyosecretome) for the Treatment of Musculoskeletal Diseases: Production Process Validation and Batch Release Test for Clinical Use" Pharmaceuticals 14, no. 6: 553. https://0-doi-org.brum.beds.ac.uk/10.3390/ph14060553









