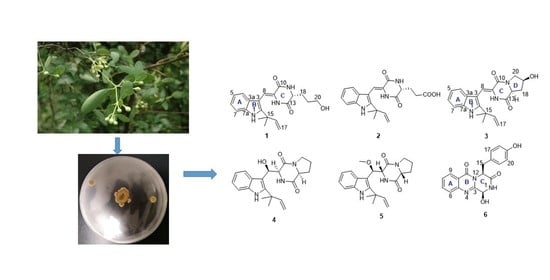
Isolation, Structural Characterization and Antidiabetic Activity of New Diketopiperazine Alkaloids from Mangrove Endophytic Fungus Aspergillus sp. 16-5c
Abstract
:1. Introduction
2. Results and Discussion
Structural Elucidation
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction, and Isolation
3.4. Antidiabetic Bioassays
3.5. ECD Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ross, S.A.; Gulve, E.A.; Wang, M. Chemistry and biochemistry of type 2 diabetes. Chem. Rev. 2004, 104, 1255–1282. [Google Scholar] [CrossRef]
- Chen, L.; Magliano, D.J.; Zimmet, P.Z. The worldwide epidemiology of type 2 diabetes mellitus-present and future perspectives. Nat. Rev. Endocrinol. 2012, 8, 228–236. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Fact Sheet: Diabetes. Available online: https://www.who.int/news-room/fact-sheets/detail/diabetes (accessed on 20 April 2020).
- Elchebly, M.; Payette, P.; Michaliszyn, E.; Cromlish, W.; Collins, S.; Loy, A.L.; Normandin, D.; Cheng, A.; Himms-Hagen, J.; Chan, C.C.; et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 1999, 283, 1544–1548. [Google Scholar] [CrossRef]
- Qian, S.; Zhang, M.; He, Y.; Wang, W.; Liu, S. Recent advances in the development of protein tyrosine phosphatase 1B inhibitors for type 2 diabetes. Fut. Med. Chem. 2016, 8, 1239–1258. [Google Scholar] [CrossRef]
- Nagaraju, K.; Ashona, S.P.; Paul, A.; Parvesh, A. Current anti-diabetic agents and their molecular targets: A review. Eur. J. Med. Chem. 2018, 152, 436–488. [Google Scholar]
- Martins, M.B.; Carvalaho, I. Diketopiperazines: Biological activity and synthesis. Tetrahedron 2007, 63, 9923–9932. [Google Scholar] [CrossRef]
- Xu, W.F.; Mao, N.; Xue, X.J.; Qi, Y.X.; Wei, M.Y.; Wang, C.Y.; Shao, C.L. Structures and absolute configurations of diketopiperazine alkaloids chrysopiperazines A–C from the gorgonian-derived Penicillium chrysogenum fungus. Mar. Drugs 2019, 17, 250. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.M.; Liang, X.A.; Kong, Y.; Jia, B. Structural diversity and biological activities of indole diketopiperazine alkaloids from fungi. J. Agric. Food Chem. 2016, 64, 6659–6671. [Google Scholar] [CrossRef]
- Wang, W.L.; Lu, Z.Y.; Tao, H.W.; Zhu, T.J.; Fang, Y.C.; Gu, Q.Q.; Zhu, W.M. Isoechinulin-type alkaloids, variecolorins A–L, from halotolerant Aspergillus wariecolor. J. Nat. Prod. 2007, 70, 1558–1564. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Kato, H.; Samizo, M.; Nojiri, Y.; Onuki, H.; Hirota, H.; Ohta, T. Notoamides F-K, prenylated Indole alkaloids isolated from a marine-derived Aspergillus sp. J. Nat. Prod. 2008, 71, 2064–2067. [Google Scholar] [CrossRef] [PubMed]
- Kuramochi, K.; Ohnishi, K.; Fujieda, S.; Nakajima, M.; Saitoh, Y.; Watanabe, N.; Takeuchi, T.; Nakazaki, A.; Sugawara, F.; Arai, T.; et al. Synthesis and biological activities of neoechinulin A derivatives: New aspects of structure–activity relationships for neoechinulin A. Chem. Pharm. Bull. 2008, 56, 1738–1743. [Google Scholar] [CrossRef] [Green Version]
- De Guzman, F.S.; Glober, J.B. New diketopiperazine metabolites from the sclerotia of Aspergillus ochrace us. J. Nat. Prod. 1992, 55, 931–939. [Google Scholar] [CrossRef]
- Cao, J.; Wang, B.-G. Chemical diversity and biological function of indolediketopiperazines from marine-derived fungi. Mar. Life Sci. Technol. 2020, 2, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.Y.; Huang, C.Y.; Guo, H.X.; Zheng, S.Y.; He, J.R.; Lin, J.; Long, Y.H. Isobenzofuranone monomer and dimer derivatives from the mangrove endophytic fungus Epicoccum nigrum SCNU-F0002 possess α-glucosidase inhibitory and antioxidant activity. Bioorg. Chem. 2019, 94, 103407. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Doi, M. Isolation of an antioxidative substance produced by Aspergillus repens. Biosci. Biotechnol. Biochem. 1999, 63, 932–933. [Google Scholar] [CrossRef]
- Gao, H.Q.; Liu, W.Z.; Zhu, T.J.; Mo, X.M.; MándIi, A.; Kurtán, T.; Li, J.; Ai, J.; Gu, Q.Q.; Li, D.H. Diketopiperazine alkaloids from a mangrove rhizosphere soil derived fungus Aspergillus effuses H1-1. Org. Biomol. Chem. 2012, 10, 9501–9506. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.L.; Cai, S.X.; Zhu, T.J.; Gu, Q.K.; Li, D.H.; Luan, Y.P. Secondarymetabolites of a deep sea derived fungus Aspergillus versicolor CXCTD-06-6a and their bioactivity. J. Ocean. Univ. Chian. 2014, 13, 691–695. [Google Scholar] [CrossRef]
- Chen, X.Q.; Si, L.L.; Liu, D.; Proksch, P.; Zhang, L.H.; Zhou, D.M.; Lin, W.H. Neoechinulin B and its analogues as potential entry inhibitors of influenza viruses, targeting viral hemagglutinin. Eur. J. Med. Chem. 2015, 93, 182–195. [Google Scholar] [CrossRef]
- Li, C.J.; Chen, P.N.; Li, H.J.; Mahmud, T.; Wu, D.L.; Xu, J.; Lan, W.J. Potential antidiabetic fumiquinazoline alkaloids from the marinederived fungus Scedosporium apiospermum F41-1. J. Nat. Prod. 2020, 83, 1082–1091. [Google Scholar] [CrossRef]
- Li, G.Y.; Yang, T.; Luo, Y.G.; Chen, X.Z.; Fang, D.M.; Zhang, G.L. Brevianamide J, a new indole alkaloid dimer from fungus Aspergillus versicolor. Org. Lett. 2009, 11, 3714–3717. [Google Scholar] [CrossRef]
- Li, G.Y.; Li, L.M.; Yang, T.; Chen, X.Z.; Fang, D.M.; Zhang, G.L. Four new alkaloids, brevianamides O–R, from the fungus Aspergillus versicolor. Helv. Chim. Acta 2010, 93, 2075–2080. [Google Scholar] [CrossRef]
- Sanz-Cervera, J.F.; Stocking, E.M.; Usui, T.; Osada, H.; Williams, R.M. Synthesis and evaluation of microtubule assembly inhibition and cytotoxicity of prenylated derivatives of cyclo-L-Trp-L-Pro. Bioorg. Med. Chem. 2000, 8, 2407–2415. [Google Scholar] [CrossRef]
- Kobayashi, M.; Aoki, S.; Gato, K.; Matsunami, K.; Kurosu, M.; Kitagawa, I. Marine natural products. XXXIV. Trisindoline, a new antibiotic indole trimer, produced by a bacterium of Vibrio sp. separated from the marine sponge Hyrtios altum. Chem. Pharm. Bull. 1994, 42, 2449–2451. [Google Scholar] [CrossRef] [Green Version]
- Sobolevskaya, M.P.; Afiyatullov, S.S.; Dyshlovoi, S.A.; Kirichuk, N.N.; Denisenko, V.A.; Kim, N.Y.; Bocharova, A.A. Metabolites from the marine isolate of the fungus Aspergillus versicolor KMM 4644. Chem. Nat. Compd. 2013, 49, 181–183. [Google Scholar] [CrossRef]
- Gubiani, J.R.; Teles, H.L.; Silva, G.H.; Young, M.C.M.; Pereira, J.O.; Bolzani, V.S.; Araujo, A.R. Cyclo-(Trp-Phe) Diketopiperazines from the endophytic fungus Aspergillus versicolor isolated from Piper aduncum. Quim. Nova 2017, 40, 138–142. [Google Scholar] [CrossRef]
- Zhong, W.M.; Wang, J.F.; Shi, X.F.; Wei, X.Y.; Chen, Y.C.; Zeng, Q.; Xiang, Y.; Chen, X.Y.; Tian, X.P.; Xiao, Z.H.; et al. Eurotiumins A–E, five new alkaloids from the marine-derived fungus Eurotium sp. SCSIO F452. Mar. Drugs 2018, 16, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.S.; Ma, K.; Tao, Q.Q.; Han, J.J.; Bao, L.; Liu, L.; Liu, H.W. Diketopiperazines and 2H-pyran-2-ones with antioxidant activity from the rice fermented with Aspergillus luchuensis. Fitoterapia 2018, 125, 266–272. [Google Scholar] [CrossRef]
- Huang, X.S.; Huang, H.B.; Li, H.X.; Sun, X.F.; Huang, H.R.; Lu, Y.J.; Lin, Y.C.; Long, Y.H.; She, Z.G. Asperterpenoid A, a new sesterterpenoid as an inhibitor of mycobacterium tuberculosis protein tyrosine phosphatase B from the culture of Aspergillus sp. 16-5c. Org. Lett. 2013, 15, 721–723. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.H.; Li, J.; Zhang, M.M.; Cui, J.; Gui, Y.H.; Zhang, Y.; Li, J.Y.; Liu, K.C.; Lin, H.W. Frondoplysins A and B, unprecedented terpene-alkaloid bioconjugates from Dysidea frondosa. Org. Lett. 2019, 21, 6190–6193. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.Q.; Ma, K.; Yang, Y.L.; Wang, K.; Chen, B.C.; Huang, Y.; Han, J.J.; Bao, L.; Liu, X.-B.; Yang, Z.L.; et al. Bioactive sesquiterpenes from the edible mushroom Flammulina velutipes and their biosynthetic pathway confirmed by genome analysis and chemical evidence. J. Org. Chem. 2016, 81, 9867–9877. [Google Scholar] [CrossRef]







| Position | 1 a | 2 a | 3 a | |||
|---|---|---|---|---|---|---|
| δC | δH (J/Hz) | δC | δH (J/Hz) | δC | δH (J/Hz) | |
| 2 | 146.2, C | 146.2, C | 146.2, C | |||
| 3 | 104.3, C | 104.3, C | 104.6, C | |||
| 3a | 127.1, C | 127.3, C | 126.0, C | |||
| 4 | 119.9, CH | 7.24, d (7.9) | 120.0, CH | 7.26, d (7.8) | 112.5, CH | 7.41, d (8.0) |
| 5 | 121.2, CH | 7.08, m | 121.2, CH | 7.08, m | 122.4, CH | 7.12, m |
| 6 | 122.5, CH | 7.13, m | 122.7, CH | 7.13, m | 121.1, CH | 7.07, m |
| 7 | 112.6, CH | 7.43, d (8.0) | 112.6, CH | 7.42, d (8.0) | 120.2, CH | 7.31, d (7.9) |
| 7a | 136.8, C | 136.9, C | 136.7, C | |||
| 8 | 114.5, CH | 7.22, s | 114.7, CH | 7.22, s | 114.6, CH | 7.23, s |
| 9 | 124.6, C | 124.5, C | 127.3, C | |||
| 10 | 162.5, C | 162.5, C | 160.9, C | |||
| 12 | 56.8, CH | 4.21, t (5.4) | 56.1, CH | 4.25, t (5.6) | 58.7, CH | 4.72, dd (11.7, 5.9) |
| 13 | 168.1, C | 167.5, C | 168.2, C | |||
| 15 | 40.5, C | 40.5, C | 40.5, C | |||
| 15a | 28.1, CH3 | 1.55, s | 28.1, CH3 | 1.55, s | 28.0, CH3 | 1.56, s |
| 15b | 28.3, CH3 | 1.54, s | 28.2, CH3 | 1.54, s | 28.3, CH3 | 1.54, s |
| 16 | 146.0, CH | 6.11, dd (17.3, 10.6) | 146.0, CH | 6.11, dd (17.3, 10.6) | 146.2, CH | 6.11, dd (17.2,10.8) |
| 17 | 112.7,CH2 | 5.11, m | 112.7, CH2 | 5.11, m | 112.5, CH2 | 5.11, m |
| 18 | 32.6, CH2 | 1.95, m | 31.0, CH2 | 2.21, m | 39.0, CH2 | 2.11, m |
| 2.03, m | 2.34, dd (13.0, 5.9) | |||||
| 19 | 28.4, CH2 | 1.67, m | 30.2, CH2 | 2.48, m | 68.6, CH | 4.51, m |
| 1.74, m | ||||||
| 20 | 62.5, CH2 | 3.63, t (6.3) | 176.3, C | 55.6, CH2 | 3.53, d (13.2) | |
| 3.92, dd
(13.2, 4.9) | ||||||
| Position | 4 a | 5 b | 6 b | |||
|---|---|---|---|---|---|---|
| δC | δH (J/Hz) | δC | δH (J/Hz) | δC | δH (J/Hz) | |
| 1 | 10.5, s | |||||
| 2 | 140.7, C | 143.6, C | 77.2, CH | 5.71, s | ||
| 3 | 111.1, C | 106.5, C | 151.4, C | |||
| 3a | 127.3, C | 128.7, C | ||||
| 4 | 110.3, CH | 7.30, d (8.0) | 112.4, CH | 7.40, d (8.1) | ||
| 5 | 118.0, CH | 6.86, m | 121.0, CH | 7.03, m | 148.4, C | |
| 6 | 120.2, CH | 6.97, m | 122.3, CH | 7.09, m | 128.2, CH | 7.73, d (8.1) |
| 7 | 121.6, CH | 7.70, d (8.0) | 121.2, CH | 7.74, d (8.0) | 136.0, CH | 7.84, ddd |
| 7a | 135.0, C | 136.6, C | (8.0, 7.0, 1.4) | |||
| 8 | 68.9, CH | 5.30, dd (3.5, 6.2) | 77.8, CH | 5.60, d (2.4) | 128.7, CH | 7.56, ddd (8.0, 7.0, 0.8) |
| 9 | 62.7, CH | 4.0, dd (4.7, 6.2) | 63.4, CH | 4.39, s | 127.6, CH | 8.14, dd (1.1, 8.0) |
| 10 | 164.4, C | 165.8, C | 121.6, C | |||
| 11 | 161.8, C | |||||
| 12 | 58.4, CH | 3.86, dd (6.9, 9.6) | 60.1, CH | 4.16, m | ||
| 13 | 169.5, C | 170.6, C | 58.9, CH | 5.37, dd (7.1, 8.4) | ||
| 14 | 8.16, d (4.5) | 171.8, C | ||||
| 15 | 38.7, C | 40.1, C | 40.4, CH2 | 3.37, dd (7.1, 13.5) | ||
| 3.44, dd (8.4, 13.5) | ||||||
| 15a | 28.1, CH3 | 1.46, s | 28.2, CH3 | 1.60, s | ||
| 15b | 28.2, CH3 | 1.44, s | 28.5, CH3 | 1.57, s | ||
| 16 | 146.3, CH | 6.16, dd (17.5, 10.5) | 146.9, CH | 6.16, dd (17.5, 10.5) | 128.4, C | |
| 17 | 110.7, CH2 | 5.09, m | 112.6, CH2 | 5.18, ddd (17.5,10.5,0.8) | 131.7, CH | 7.06, d (8.4) |
| 18 | 28.8, CH2 | 2.09, m | 30.0, CH2 | 1.95, m | 116.1, CH | 6.64, d (8.4) |
| 1.73, m | 2.34, dd (7.9, 3.4) | |||||
| 19 | 21.9, CH2 | 1.83, m | 22.9, CH2 | 1.95, m | 157.6, C | |
| 2.06, m | ||||||
| 20 | 45.3, CH2 | 3.26, m | 46.1, CH2 | 3.47, m | 116.1, CH | 6.64, d (8.4) |
| 3.40, m | 3.76, m | |||||
| 21 | 131.7, CH | 7.06, d (8.4) | ||||
| 8-OH | 5.33, d (3.5) | |||||
| 8-OCH3 | 56.9, CH3 | 3.17, s | ||||
| Compd. | α-glucosidase
(IC50/μM) | PTP1B (Inhibition Ratio/%) a | Compd. | α-glucosidase (IC50/μM) | PTP1B (Inhibition Ratio/%) a |
|---|---|---|---|---|---|
| 1 | 18.2 | 29 | 10 | 40.7 | <10 |
| 2 | 130.7 | <10 | 11 | 37.5 | <10 |
| 3 | 83.9 | <10 | 12 | 1086.6 | <10 |
| 4 | 144.2 | <10 | 13 | 480.5 | <10 |
| 5 | 1093.5 | <10 | 14 | 353.2 | <10 |
| 6 | 267.3 | <10 | 15 | 67.8 | <10 |
| 7 | 198.2 | <10 | 16 | 362.6 | <10 |
| 8 | 364.3 | <10 | Acarbose b | 408.0 | - |
| 9 | 7.6 | <10 | Oleanolic acid c | - | 99.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, G.; Huang, C.; Li, J.; Chen, T.; Tang, J.; Liu, W.; Long, Y. Isolation, Structural Characterization and Antidiabetic Activity of New Diketopiperazine Alkaloids from Mangrove Endophytic Fungus Aspergillus sp. 16-5c. Mar. Drugs 2021, 19, 402. https://0-doi-org.brum.beds.ac.uk/10.3390/md19070402
Ye G, Huang C, Li J, Chen T, Tang J, Liu W, Long Y. Isolation, Structural Characterization and Antidiabetic Activity of New Diketopiperazine Alkaloids from Mangrove Endophytic Fungus Aspergillus sp. 16-5c. Marine Drugs. 2021; 19(7):402. https://0-doi-org.brum.beds.ac.uk/10.3390/md19070402
Chicago/Turabian StyleYe, Geting, Cuiying Huang, Jialin Li, Tao Chen, Jing Tang, Wenbin Liu, and Yuhua Long. 2021. "Isolation, Structural Characterization and Antidiabetic Activity of New Diketopiperazine Alkaloids from Mangrove Endophytic Fungus Aspergillus sp. 16-5c" Marine Drugs 19, no. 7: 402. https://0-doi-org.brum.beds.ac.uk/10.3390/md19070402