Tannery Wastewater Recalcitrant Compounds Foster the Selection of Fungi in Non-Sterile Conditions: A Pilot Scale Long-Term Test
Abstract
:1. Introduction
2. Materials and Methods
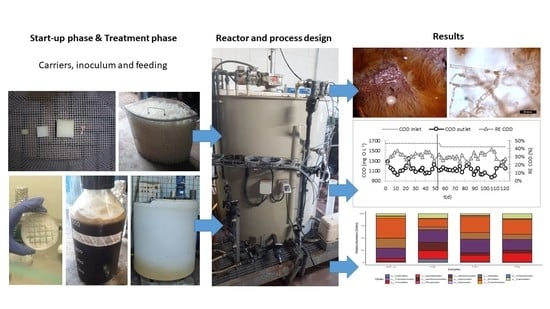
2.1. Inoculum Preparation
2.2. Experimental Set-Up
2.3. Process Operation: Start-Up
2.4. Process Operation: Treatment Phase
2.5. Analytical Methods and Statistical Analysis
2.6. Metagenomic DNA Extraction
2.7. Metabarcoding of the Fungal and Bacterial Biodiversity
3. Results
3.1. Start-Up Phase
3.2. Treatment Phase
3.3. Microbial Taxa Distribution Changes along the Process
3.4. Bacterial Diversity Indexes and Core Microbiome
4. Discussion
4.1. Reactor Performance and Advanced Application
4.2. Structure Community Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meffe, R.; de Bustamante, I. Emerging organic contaminants in surface water and groundwater: A first overview of the situation in Italy. Sci. Total Environ. 2014, 481, 280–295. [Google Scholar] [CrossRef]
- Liu, Z.; Kanjo, Y.; Mizutani, S. Removal mechanisms for endocrine disrupting compounds (EDCs) in wastewater treatment—physical means, biodegradation, and chemical advanced oxidation: A review. Sci. Total Environ. 2009, 407, 731–748. [Google Scholar] [CrossRef] [PubMed]
- Li, W.C. Occurrence, sources, and fate of pharmaceuticals in aquatic environment and soil. Environ. Pollut. 2014, 187, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Guo, W.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I.; Zhang, J.; Liang, S.; Wang, X.C. A review on the occurrence of micropollutants in the aquatic environment and their fate and removal during wastewater treatment. Sci. Total Environ. 2014, 473–474, 619–641. [Google Scholar] [CrossRef] [PubMed]
- Bui, X.T.; Vo, T.P.T.; Ngo, H.H.; Guo, W.S.; Nguyena, T.T. Multicriteria assessment of advanced treatment technologies for micropollutants removal at large-scale applications. Sci. Total Environ. 2016, 563, 1050–1067. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.O.; Moreira, N.F.F.; Ribeiro, A.R.; Pereira, M.F.R.; Silva, A.M.T. Occurrence and removal of organic micropollutants: An overview of the watch list of EU Decision 2015/495. Water Res. 2016, 94, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef]
- Svobodová, K.; Novotný, C. Bioreactors based on immobilized fungi: Bioremediation under non-sterile conditions. Biotechnol. Appl. Microbiol. 2018, 102, 39–46. [Google Scholar] [CrossRef]
- Espinosa-Ortiz, E.J.; Rene, E.R.; Pakshirajan, K.; van Hullebusch, E.D.; Lens, P.N.L. Fungal pelleted reactors in wastewater treatment: Applications and perspectives. Chem. Eng. J. 2016, 283, 553–571. [Google Scholar] [CrossRef]
- Rene, E.R.; Veiga, M.C.; Kennes, C. Biodegradation of gas-phase styrene using the fungus Sporothrix variecibatus: Impact of pollutant load and transient operation. Chemosphere 2010, 79, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Gao, D.; Zeng, Y.; Wen, X.; Qian, Y. Competition strategies for the incubation of white rot fungi under non-sterile conditions. Process Biochem. 2008, 43, 937–944. [Google Scholar] [CrossRef]
- Tigini, V.; Bevione, F.; Prigione, V.; Poli, A.; Ranieri, L.; Spennati, F.; Munz, G.; Varese, G.C. Tannery mixed liquors from an ecotoxicological and mycological point of view: Risks vs. potential biodegradation application. Sci. Total Environ. 2018, 627, 835–843. [Google Scholar] [CrossRef]
- Tigini, V.; Bevione, F.; Prigione, V.; Poli, A.; Ranieri, L.; Spennati, F.; Munz, G.; Varese, G.C. Wastewater-Agar as a selection environment: A first step towards a fungal in-situ bioaugmentation strategy. Ecotoxicol. Environ. Saf. 2019, 171, 443–450. [Google Scholar] [CrossRef]
- Lorenz, M.M.; Alkhafadji, L.; Stringano, E.; Nilsson, S.; Mueller-Harvey, I.; Udén, P. Relationship between condensed tannin structures and their ability to precipitate feed proteins in the rumen. J. Sci. Food Agric. 2014, 94, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Mannucci, A.; Munz, G.; Mori, G.; Lubello, C. Anaerobic treatment of vegetable tannery wastewaters: A review. Desalination 2010, 264, 1–8. [Google Scholar] [CrossRef]
- Spennati, F.; Ricotti, A.; Mori, G.; Siracusa, G.; Becarelli, S.; Di Gregorio, S.; Tigini, V.; Varese, G.C.; Munz, G. The role of cosubstrate and mixing on fungal biofilm efficiency in the removal of tannins. Environ. Technol. 2020, 41, 3515–3523. [Google Scholar] [CrossRef] [PubMed]
- Bardi, A.; Yuan, Q.; Siracusa, G.; Chicca, I.; Islam, M.; Spennati, F.; Tigini, V.; Di Gregorio, S.; Levin, D.B.; Petroni, G.; et al. Effect of cellulose as co-substrate on old landfill leachate treatment using white-rot fungi. Bioresour. Technol. 2017, 241, 1067–1076. [Google Scholar] [CrossRef]
- Spennati, F.; Mora, M.; Tigini, V.; La China, S.; Di Gregorio, S.; Gabriel, D.; Munz, G. Removal of Quebracho and Tara tannins in fungal bioreactors: Performance and biofilm stability analysis. J. Environ. Manag. 2019, 231, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Armitage, D.W.; Gallagher, K.L.; Youngblut, N.D.; Buckley, D.H.; Zinder, S.H. Millimeter-scale patterns of phylogenetic and trait diversity in a salt marsh microbial mat. Front. Microbiol. 2012, 3, 293. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Shetty, S.A.; Hugenholtz, F.; Lahti, L.; Smidt, H.; de Vos, W.M. Intestinal microbiome landscaping: Insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol. Rev. 2017, 41, 182–199. [Google Scholar] [CrossRef] [PubMed]
- Spennati, F.; Mora, M.; Bardi, A.; Becarelli, S.; Siracusa, G.; Di Gregorio, S.; Gabriel, D.; Mori, G.; Munz, G. Respirometric techniques coupled with lab-scale tests for kinetic and stoichiometric characterisation of fungal and bacterial tannin-degrading biofilms. Water Sci. Technol. 2020, 81, 2559–2567. [Google Scholar] [CrossRef]
- Djelal, H.; Amrane, A. Biodegradation by bioaugmentation of dairy wastewater by fungal consortium on a bioreactor lab-scale and on a pilot-scale. J. Environ. Sci. 2013, 25, 1906–1912. [Google Scholar] [CrossRef] [Green Version]
- Acikgoz, C.; Gül, Ü.D.; Özan, K.; Borazan, A.A. Degradation of Reactive Blue by the mixed culture of Aspergillus versicolor and Rhizopus arrhizus in membrane bioreactor (MBR) system. Desalin. Water Treat. 2016, 57, 3750–3756. [Google Scholar] [CrossRef]
- Křesinová, Z.; Linhartová, L.; Filipová, A.; Ezechiáš, M.; Mašín, P.; Cajthaml, T. Biodegradation of endocrine disruptors in urban wastewater using Pleurotus ostreatus bioreactor. New Biotechnol. 2016, 43, 53–61. [Google Scholar] [CrossRef]
- Prigione, V.; Spina, F.; Tigini, V.; Giovando, S.; Varese, G.C. Biotransformation of industrial tannins by filamentous fungi. Appl. Microbiol. Biotechnol. 2018, 102, 10361–10375. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Lee, S.; Lee, S.H.; Kim, S.R.; Oh, H.-S.; Park, P.-K.; Choo, K.-H.; Kim, Y.-W.; Lee, J.-K.; Lee, C.-H. Fungal Quorum Quenching: A Paradigm Shift for Energy Savings in Membrane Bioreactor (MBR) for Wastewater Treatment. Environ. Sci. Technol. 2016, 50, 10914–10922. [Google Scholar] [CrossRef] [PubMed]
- Bellotti, N.; Deyá, C.; Del Amo, B.; Romagnoli, R. “Quebracho” tannin derivative and boosters biocides for new antifouling formulations. J. Coat. Technol. Res. 2012, 9, 551–559. [Google Scholar] [CrossRef]
- Lofrano, G.; Meriç, S.; Zengin, G.E.; Orhon, D. Chemical and biological treatment technologies for leather tannery chemicals and wastewaters: A review. Sci. Total Environ. 2013, 461, 265–281. [Google Scholar] [CrossRef]
- Romero-Dondiz, E.M.; Almazán, J.E.; Rajal, V.B.; Castro-Vidaurre, E.F. Comparison of the performance of ultrafiltration and nanofiltration membranes for recovery and recycle of tannins in the leather industry. J. Clean. Prod. 2016, 135, 71–79. [Google Scholar] [CrossRef]
- Hassoune, J.; Tahiri, S.; Aarfane, A.; El krati, M. Removal of Hydrolyzable and Condensed Tannins from Aqueous Solutions by Electrocoagulation Process. J. Environ. Eng. 2017, 143, 04017010. [Google Scholar] [CrossRef]
- Ramírez-Ramírez, N.; Espinosa-Lloréns, M.; Fernández-García, L.A.; Véliz-Lorenzo, E.; Ramos-Rodríguez, Y. Tratamiento con ozono de agua residual con taninos de curtiduría al vegetal/Treatment with Ozone of Wastewater Containing Tannins from Vegetal Tannery. Tecnol. Cienc. Agua. 2016, 7, 53–73. [Google Scholar]
- Liu, J.; Chen, H.; Xu, Z.; Zheng, S.; Xue, M. Adsorption of tannic acid from aqueous solution by aminopropyl functionalized SBA-15. Desalin. Water Treat. 2015, 56, 475–484. [Google Scholar] [CrossRef]
- Cokgor, E.U.; Karahan, O.; Orhon, D. The effect of mixing pharmaceutical and tannery wastewaters on the biodegradation characteristics of the effluents. J. Hazard. Mater. 2008, 156, 292–299. [Google Scholar] [CrossRef]
- Becarelli, S.; Chicca, I.; Siracusa, G.; La China, S.; Gentini, A.; Lorenzi, R.; Munz, G.; Petroni, G.; Levin, D.B.; Di Gregorio, S. Hydrocarbonoclastic Ascomycetes to enhance co-composting of total petroleum hydrocarbon (TPH) contaminated dredged sediments and lignocellulosic matrices. New Biotechnol. 2019, 50, 27–36. [Google Scholar] [CrossRef]
- Wrede, D.; Taha, M.; Miranda, A.F.; Kadali, K.; Stevenson, T.; Ball, A.S.; Mouradov, A. Co-cultivation of fungal and microalgal cells as an efficient system for harvesting microalgal cells, lipid production and wastewater treatment. PLoS ONE 2014, 9, e113497. [Google Scholar] [CrossRef] [Green Version]
- Kanniah Goud, R.; Arunasri, K.; Yeruva, D.K.; Krishna, K.V.; Dahiya, S.; Mohan, S.V. Impact of selectively enriched microbial communities on long-term fermentative biohydrogen production. Bioresour. Technol. 2017, 242, 253–264. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, A.; Osorio, F.; Morillo, J.A.; Rodriguez-Sanchez, A.; Gonzalez-Lopez, J.; Abbas, B.A.; van Loosdrecht, M.C.M. Comparison of bacterial diversity in full scale anammox bioreactors operated under different conditions. Biotechnol. Prog. 2015, 31, 1464–1472. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Tolalpa, L.; Jiménez, D.J.; de Lima Brossi, M.J.; Salles, J.F.; van Elsas, J.D. Different inocula produce distinctive microbial consortia with similar lignocellulose degradation capacity. Appl. Microbiol. Biotechnol. 2016, 100, 7713–7725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Ge, J.; Wang, S.; Su, H. Insight into formation and biological characteristics of Aspergillus tubingensis-based aerobic granular sludge (AT-AGS) in wastewater treatment. Sci. Total Environ. 2020, 739, 140128. [Google Scholar] [CrossRef]
- Nasir, N.M.; Bakar, N.S.A.; Lananan, F.; Hamid, F.H.A.; Lam, S.S.; Jusoh, A. Treatment of African catfish, Clarias gariepinus wastewater utilizing phytoremediation of microalgae, Chlorella sp. with Aspergillus niger bio-harvesting. Bioresour. Technol. 2015, 190, 492–498. [Google Scholar] [CrossRef]
- Kazartsev, I.A.; Serova, T.A.; Titova, Y.A.; Gannibal, P.B. Identification of wood-inhabiting fungal communities in two historical buildings of St. Petersburg. Mikol. Fitopatol. 2014, 48, 172–181. [Google Scholar]
- Okafor, N. Environmental Microbiology of Aquatic and Waste Systems; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar]
- Świkatczak, P.; Cydzik-Kwiatkowska, A. Performance and microbial characteristics of biomass in a full-scale aerobic granular sludge wastewater treatment plant. Environ. Sci. Pollut. Res. 2018, 25, 1655–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, C.M.; Liu, W.-T. Community structure analysis of reverse osmosis membrane biofilms and the significance of Rhizobiales bacteria in biofouling. Environ. Sci. Technol. 2007, 41, 4728–4734. [Google Scholar] [CrossRef] [PubMed]
- Erlacher, A.; Cernava, T.; Cardinale, M.; Soh, J.; Sensen, C.W.; Grube, M.; Berg, G. Rhizobiales as functional and endosymbiontic members in the lichen symbiosis of Lobaria pulmonaria L. Front. Microbiol. 2015, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-I.; Kim, J.-O.; Lee, Y.-R.; Ekpeghere, K.I.; Koh, S.-C.; Whang, K.-S. Denitratimonas tolerans gen. nov., sp. nov., a denitrifying bacterium isolated from a bioreactor for tannery wastewater treatment. Antonie Leeuwenhoek 2016, 109, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Shao, M.-F.; Ye, L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012, 6, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Spennati, F.; Mori, G.; Munz, G.; Vannini, C. The microbial community in a moving bed biotrickling filter operated to remove hydrogen sulfide from gas streams. Syst. Appl. Microbiol. 2018, 41, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Feng, J.; Chen, S.; Li, B.; Sekar, R.; Zhao, Z.; Jia, J.; Wang, Y.; Kang, P. Disentangling the drivers of diversity and distribution of fungal community composition in wastewater treatment plants across spatial scales. Front. Microbiol. 2018, 9, 1291. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spennati, F.; La China, S.; Siracusa, G.; Di Gregorio, S.; Bardi, A.; Tigini, V.; Mori, G.; Gabriel, D.; Munz, G. Tannery Wastewater Recalcitrant Compounds Foster the Selection of Fungi in Non-Sterile Conditions: A Pilot Scale Long-Term Test. Int. J. Environ. Res. Public Health 2021, 18, 6348. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph18126348
Spennati F, La China S, Siracusa G, Di Gregorio S, Bardi A, Tigini V, Mori G, Gabriel D, Munz G. Tannery Wastewater Recalcitrant Compounds Foster the Selection of Fungi in Non-Sterile Conditions: A Pilot Scale Long-Term Test. International Journal of Environmental Research and Public Health. 2021; 18(12):6348. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph18126348
Chicago/Turabian StyleSpennati, Francesco, Salvatore La China, Giovanna Siracusa, Simona Di Gregorio, Alessandra Bardi, Valeria Tigini, Gualtiero Mori, David Gabriel, and Giulio Munz. 2021. "Tannery Wastewater Recalcitrant Compounds Foster the Selection of Fungi in Non-Sterile Conditions: A Pilot Scale Long-Term Test" International Journal of Environmental Research and Public Health 18, no. 12: 6348. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph18126348