Mode of Transmission Determines the Virulence of Black Queen Cell Virus in Adult Honey Bees, Posing a Future Threat to Bees and Apiculture
Abstract
:1. Introduction
2. Materials and Methods
2.1. Honey Bees
2.2. BQCV Inoculation
2.3. RNA Extraction and Detection of Virus
2.4. Gene Expression
2.5. Statistical Analysis
3. Results
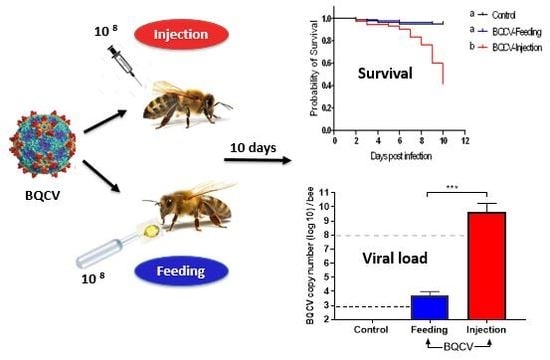
3.1. Virulence
3.2. Effects on Viral Titer
3.3. Effects on Gene Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Viral Target | Primer Name | Sequence | qPCR Efficiency | Reference |
|---|---|---|---|---|
| ABPV * | KIABPV-F6648 | CCTTTCATGATGTGGAAAC | --- | [64] |
| KIABPV-B6707 | CTGAATAATACTGTGCGTATC | |||
| BQCV | BQCV-qF7893 | AGTGGCGGAGATGTATGC | 103.3% | [65] |
| BQCV-qB8150 | GGAGGTGAAGTGGCTATATC | |||
| CBPV | CBPV1-qF1818 | CAACCTGCCTCAACACAG | 90.2% | [65] |
| CBPV1-qB2077 | AATCTGGCAAGGTTGACTGG | |||
| DWV-A | DWV-F8668 | TTCATTAAAGCCACCTGGAACATC | 85.6% | [66] |
| DWV-B8757 | TTTCCTCATTAACTGTGTCGTTGA | |||
| DWV-B | VDVq-F2 | TAT CTT CAT TAA AAC CGC CAG GCT | 86.0% | [32] |
| VDVq-R2a | CTT CCT CAT TAA CTG AGT TGT TGTC | |||
| SBPV | SBPV-F3177 | GCGCTTTAGTTCAATTGCC | 93.8% | [67] |
| SBPV-B3363 | ATTATAGGACGTGAAAATATAC | |||
| SBV | SBV-qF3164 | TTGGAACTACGCATTCTCTG | 92.1% | [65] |
| SBV-qB3461 | GCTCTAACCTCGCATCAAC |
| Locus | Category | Sequence | qPCR Efficiency | Reference/Accession Number |
|---|---|---|---|---|
| Dicer-like | Innate immunity | F: CCAACAGGAGCTGGAAAAAC | 103% | [31] XM_006571316.1 |
| R: TCTCCACTAAGTGCTGCACAA | ||||
| Argonaute 2 (AGO2) | Innate immunity | F:TCAACAGCAGCAATCGGATA | 104% | [31] XM_395048.5 |
| R:TTGCGGTGAACTTTGTTGTT | ||||
| Tarbp2-like | Innate immunity | F: AGGGTTTGCCACATGAAAGA | 102% | [31] XM_006564411.1 |
| R: AATCAGCATAACGGGCACTC | ||||
| AMActin (β-actin) | Housekeeping | F: ATGCCAACACTGTCCTTTCTGG | 101% | [66] |
| R: GACCCACCAATCCATACGGA |
References
- Hung, K.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B Biol. Sci. 2018, 285. [Google Scholar] [CrossRef] [Green Version]
- Al Naggar, Y.; Codling, G.; Giesy, J.P.; Safer, A. Beekeeping and the need for pollination from an agricultural perspective in Egypt. Bee World 2018, 95, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Brutscher, L.M.; McMenamin, A.J.; Flenniken, M.L. The buzz about honey bee viruses. PLoS Pathog. 2016, 12, e1005757. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, A.J.; Flenniken, M.L. Recently identified bee viruses and their impact on bee pollinators. Curr. Opin. Insect Sci. 2018, 26, 120–129. [Google Scholar] [CrossRef]
- McMenamin, A.J.; Genersch, E. Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 2015, 8, 121–129. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Flenniken, M.L. Bee viruses: Ecology, pathogenicity, and impacts. Annu. Rev. Èntomol. 2019, 64, 205–226. [Google Scholar] [CrossRef]
- Schmid-Hempel, P. Evolutionary Parasitology; Oxford University Press (OUP): Oxford, UK, 2011. [Google Scholar]
- Cressler, C.E.; McLeod, D.; Rozins, C.; Hoogen, J.V.D.; Day, T. The adaptive evolution of virulence: A review of theoretical predictions and empirical tests. Parasitology 2015, 143, 915–930. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Evans, J.D.; Feldlaufer, M. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebr. Pathol. 2006, 92, 152–159. [Google Scholar] [CrossRef]
- Le Clec’H, W.; Dittmer, J.; Raimond, M.; Bouchon, D.; Sicard, M. Phenotypic shift in Wolbachia virulence towards its native host across serial horizontal passages. Proc. R. Soc. B Biol. Sci. 2017, 284. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y. Viruses and viral diseases of the honey bee, Apis mellifera. In Recent Advances in Entomological Research; Springer: Berlin, Germany, 2011; pp. 105–120. [Google Scholar] [CrossRef]
- Ewald, P.W. Host-parasite relations, vectors, and the evolution of disease severity. Annu. Rev. Ecol. Syst. 1983, 14, 465–485. [Google Scholar] [CrossRef]
- Froissart, R.; Doumayrou, J.; Vuillaume, F.; Alizon, S.; Michalakis, Y. The virulence–transmission trade-off in vector-borne plant viruses: A review of (non-)existing studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1907–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, T. The evolution of virulence in vector-borne and directly transmitted parasites. Theor. Popul. Biol. 2002, 62, 199–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, D.P.; Wilfert, L.; Paxton, R.J.; Brown, M.J.F. Emerging viruses in bees: From molecules to ecology. In Advances in Applied Microbiology; Elsevier: Cambridge, MA, USA, 2018; pp. 251–291. [Google Scholar]
- Nazzi, F.; Le Conte, Y. Ecology of Varroa destructor, the major ectoparasite of the Western honey bee, Apis mellifera. Annu. Rev. Èntomol. 2016, 61, 417–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erban, T.; Harant, K.; Hubalek, M.; Vitamvas, P.; Kamler, M.; Poltronieri, P.; Tyl, J.; Markovic, M.; Titera, D. In-depth proteomic analysis of Varroa destructor: Detection of DWV-complex, ABPV, VdMLV and honeybee proteins in the mite. Sci. Rep. 2015, 5, 13907. [Google Scholar] [CrossRef] [Green Version]
- Ramsey, S.; Ochoa, R.; Bauchan, G.R.; Gulbronson, C.; Mowery, J.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792–1801. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.J.; Brettell, L. Deformed wing virus in honeybees and other insects. Annu. Rev. Virol. 2019, 6, 49–69. [Google Scholar] [CrossRef]
- Martin, S.J.; Highfield, A.C.; Brettell, L.; Villalobos, E.M.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global honey bee viral landscape altered by a parasitic mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef]
- Mondet, F.; De Miranda, J.R.; Kretzschmar, A.; Le Conte, Y.; Mercer, A. On the front line: Quantitative virus dynamics in honeybee (Apis mellifera L.) colonies along a new expansion front of the parasite Varroa destructor. PLoS Pathog. 2014, 10, e1004323. [Google Scholar] [CrossRef] [Green Version]
- Crailsheim, K.; Riessberger-Gallé, U. Honey bee age-dependent resistance against American Foulbrood. Apidologie 2001, 32, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; Spivak, M. Socialized medicine: Individual and communal disease barriers in honey bees. J. Invertebr. Pathol. 2010, 103, S62–S72. [Google Scholar] [CrossRef]
- Kingsolver, M.B.; Huang, Z.; Hardy, R.W. Insect antiviral innate immunity: Pathways, effectors, and connections. J. Mol. Biol. 2013, 425, 4921–4936. [Google Scholar] [CrossRef] [Green Version]
- Merkling, S.H.; Van Rij, R.P. Beyond RNAi: Antiviral defense strategies in Drosophila and mosquito. J. Insect Physiol. 2013, 59, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Cherry, S. Viruses and antiviral immunity in Drosophila. Dev. Comp. Immunol. 2013, 42, 67–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemp, C.; Mueller, S.; Goto, A.; Barbier, V.; Paro, S.; Bonnay, F.; Dostert, C.; Troxler, L.; Hetru, C.; Meignin, C.; et al. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 2012, 190, 650–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nayak, A.; Tassetto, M.; Kunitomi, M.; Andino, R. RNA Interference-mediated intrinsic antiviral immunity in invertebrates. Curr. Top. Microbiol. Immunol. 2013, 371, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.W.; Van Rij, R.P. The long and short of antiviral defense: Small RNA-based immunity in insects. Curr. Opin. Virol. 2014, 7, 19–28. [Google Scholar] [CrossRef]
- Karlikow, M.; Goic, B.; Saleh, M.-C. RNAi and antiviral defense in Drosophila: Setting up a systemic immune response. Dev. Comp. Immunol. 2014, 42, 85–92. [Google Scholar] [CrossRef] [Green Version]
- Galbraith, D.A.; Yang, X.; Nino, E.L.; Yi, S.; Grozinger, C. Parallel epigenomic and transcriptomic responses to viral infection in honey bees (Apis mellifera). PLoS Pathog. 2015, 11, e1004713. [Google Scholar] [CrossRef] [Green Version]
- McMahon, D.P.; Fürst, M.; Caspar, J.; Theodorou, P.; Brown, M.J.F.; Paxton, R.J. A sting in the spit: Widespread cross-infection of multiple RNA viruses across wild and managed bees. J. Anim. Ecol. 2015, 84, 615–624. [Google Scholar] [CrossRef]
- Radzeviciute, R.; Theodorou, P.; Husemann, M.; Japoshvili, G.; Kirkitadze, G.; Zhusupbaeva, A.; Paxton, R.J. Replication of honey bee-associated RNA viruses across multiple bee species in apple orchards of Georgia, Germany and Kyrgyzstan. J. Invertebr. Pathol. 2017, 146, 14–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alger, S.A.; Burnham, P.A.; Brody, A.K. Flowers as viral hot spots: Honey bees (Apis mellifera) unevenly deposit viruses across plant species. PLoS ONE 2019, 14, e0221800. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L.; Woods, R.D. Two More Small RNA viruses from honey bees and further observations on Sacbrood and Acute bee-paralysis viruses. J. Gen. Virol. 1977, 37, 175–182. [Google Scholar] [CrossRef]
- Leat, N.; Ball, B.; Govan, V.; Davison, S. Analysis of the complete genome sequence of Black queen-cell virus, a picorna-like virus of honey bees. J. Gen. Virol. 2000, 81, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- Spurny, R.; Přidal, A.; Pálková, L.; Kiem, H.K.T.; De Miranda, J.R.; Plevka, P. Virion structure of Black queen cell virus, a common honeybee pathogen. J. Virol. 2017, 91, e02100-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, M.; Ball, B. The incidence and world distribution of honey bee viruses. Bee World 1996, 77, 141–162. [Google Scholar] [CrossRef]
- Bailey, L.; Ball, B.V.; Perry, J.N. Association of viruses with two protozoal pathogens of the honey bee. Ann. Appl. Biol. 1983, 103, 13–20. [Google Scholar] [CrossRef]
- Doublet, V.; Labarussias, M.; De Miranda, J.R.; Moritz, R.; Paxton, R.J. Bees under stress: Sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ. Microbiol. 2014, 17, 969–983. [Google Scholar] [CrossRef]
- Doublet, V.; Paxton, R.J.; McDonnell, C.M.; Dubois, E.; Nidelet, S.; Moritz, R.F.; Alaux, C.; Le Conte, Y. Brain transcriptomes of honey bees (Apis mellifera) experimentally infected by two pathogens: Black queen cell virus and Nosema ceranae. Genom. Data 2016, 10, 79–82. [Google Scholar] [CrossRef]
- Tehel, A.; Quynh, V.; Bigot, D.; Gogol-Döring, A.; Koch, P.; Jenkins, C.; Doublet, V.; Theodorou, P.; Paxton, R.J. The two prevalent genotypes of an emerging infectious disease, Deformed wing virus, cause equally low pupal mortality and equally high wing deformities in host honey bees. Viruses 2019, 11, 114. [Google Scholar] [CrossRef] [Green Version]
- De Miranda, J.R.; Bailey, L.; Ball, B.V.; Blanchard, P.E.; Budge, G.; Chejanovsky, N.; Chen, Y.-P.; Gauthier, L.; Genersch, E.; De Graaf, D.C.; et al. Standard methods for virus research in Apis mellifera. J. Apic. Res. 2013, 52, 1–56. [Google Scholar] [CrossRef] [Green Version]
- Carrillo-Tripp, J.; Dolezal, A.G.; Goblirsch, M.J.; Miller, W.A.; Toth, A.L.; Bonning, B.C. In vivo and in vitro infection dynamics of honey bee viruses. Sci. Rep. 2016, 6, 22265. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.R.; Alaux, C.; Costa, C.; Csáki, T.; Doublet, V.; Eisenhardt, D.; Fries, I.; Kuhn, R.; McMahon, D.P.; Medrzycki, P.; et al. Standard methods for maintaining adult Apis mellifera in cages under in vitro laboratory conditions. J. Apic. Res. 2013, 52, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Antiviral defense mechanisms in honey bees. Curr. Opin. Insect Sci. 2015, 10, 71–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, P. Q-Gene: Processing quantitative real-time RT-PCR data. Bioinformatics 2003, 19, 1439–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Möckel, N.; Gisder, S.; Genersch, E. Horizontal transmission of Deformed wing virus: Pathological consequences in adult bees (Apis mellifera) depend on the transmission route. J. Gen. Virol. 2010, 92, 370–377. [Google Scholar] [CrossRef]
- Jousset, F.X.; Plus, N.; Croizier, G.; Thomas, M. Existence chez Drosophila de deux groupes de Picomavirus de propriétés sérologiques et biologiques différentes. C. R. Acad. Sci. 1972, 275, 3043–3046. [Google Scholar]
- Thomas-Orillard, M. Modifications of mean ovariole number, fresh weight of adult females and developmental time in Drosophila melanogaster induced by Drosophila C virus. Genetics 1984, 107, 635–644. [Google Scholar]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Genet. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Engel, P.; Kwong, W.K.; Mcfrederick, Q.; Anderson, K.E.; Barribeau, S.M.; Chandler, J.A.; Cornman, R.S.; Dainat, J.; De Miranda, J.R.; Doublet, V.; et al. The bee microbiome: Impact on bee health and model for evolution and ecology of host-microbe interactions. MBio 2016, 7, e02164-15. [Google Scholar] [CrossRef] [Green Version]
- Bailey, L.; Woods, R.D. Three previously undescribed viruses from the honey bee. J. Gen. Virol. 1974, 25, 175–186. [Google Scholar] [CrossRef]
- Remnant, E.J.; Mather, N.; Gillard, T.L.; Yagound, B.; Beekman, M. Direct transmission by injection affects competition among RNA viruses in honeybees. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182452. [Google Scholar] [CrossRef] [PubMed]
- Berényi, O.; Bakonyi, T.; Derakhshifar, I.; Koglberger, H.; Nowotny, N. Occurrence of six honeybee viruses in diseased Austrian apiaries. Appl. Environ. Microbiol. 2006, 72, 2414–2420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tentcheva, D.; Gauthier, L.; Zappulla, N.; Dainat, B.; Cousserans, F.; Colin, M.E.; Bergoin, M. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 2004, 70, 7185–7191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, J.M.K.; Anderson, D.L.; Durr, P.A. Absence of Deformed wing virus and Varroa destructor in Australia provides unique perspectives on honeybee viral landscapes and colony losses. Sci. Rep. 2017, 7, 6925. [Google Scholar] [CrossRef] [PubMed]
- Hatjina, F.; Tsoktouridis, G.; Bouga, M.; Charistos, L.; Evangelou, V.; Avtzis, D.N.; Meeus, I.; Brunain, M.; Smagghe, G.; De Graaf, D.C. Polar tube protein gene diversity among Nosema ceranae strains derived from a Greek honey bee health study. J. Invertebr. Pathol. 2011, 108, 131–134. [Google Scholar] [CrossRef]
- Alger, S.A.; Burnham, P.A.; Boncristiani, H.F.; Brody, A.K. RNA virus spillover from managed honeybees (Apis mellifera) to wild bumblebees (Bombus spp.). PLoS ONE 2019, 14, e0217822. [Google Scholar] [CrossRef] [Green Version]
- Natsopoulou, M.E.; McMahon, D.P.; Doublet, V.; Frey, E.; Rosenkranz, P.; Paxton, R.J. The virulent, emerging genotype B of Deformed wing virus is closely linked to overwinter honeybee worker loss. Sci. Rep. 2017, 7, 5242. [Google Scholar] [CrossRef]
- Gammon, D.B.; Mello, C.C. RNA interference-mediated antiviral defense in insects. Curr. Opin. Insect Sci. 2015, 8, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Aliyari, R.; Wu, Q.; Li, H.-W.; Wang, X.-H.; Li, F.; Green, L.D.; Han, C.; Li, W.-X.; Ding, S. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 2008, 4, 387–397. [Google Scholar] [CrossRef] [Green Version]
- De Smet, L.; Ravoet, J.; Wenseleers, T.; De Graaf, D.C. Expression of key components of the RNAi machinery are suppressed in Apis mellifera that suffer a high virus infection. Èntomol. Sci. 2017, 20, 76–85. [Google Scholar] [CrossRef] [Green Version]
- De Miranda, J.R.; Cordoni, G.; Budge, G. The Acute bee paralysis virus–Kashmir bee virus–Israeli acute paralysis virus complex. J. Invertebr. Pathol. 2010, 103, S30–S47. [Google Scholar] [CrossRef] [PubMed]
- Locke, B.; Le Conte, Y.; Crauser, D.; Fries, I. Host adaptations reduce the reproductive success of Varroa destructor in two distinct European honey bee populations. Ecol. Evol. 2012, 2, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Forsgren, E.; De Miranda, J.R.; Isaksson, M.; Wei, S.; Fries, I. Deformed wing virus associated with Tropilaelaps mercedesae infesting European honey bees (Apis mellifera). Exp. Appl. Acarol. 2008, 47, 87–97. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.R.; Dainat, B.; Locke, B.; Cordoni, G.; Berthoud, H.; Gauthier, L.; Neumann, P.; Budge, G.E.; Ball, B.V.; Stoltz, D.B. Genetic characterization of Slow bee paralysis virus of the honeybee (Apis mellifera L.). J. Gen. Virol. 2010, 91, 2524–2530. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Naggar, Y.; Paxton, R.J. Mode of Transmission Determines the Virulence of Black Queen Cell Virus in Adult Honey Bees, Posing a Future Threat to Bees and Apiculture. Viruses 2020, 12, 535. https://0-doi-org.brum.beds.ac.uk/10.3390/v12050535
Al Naggar Y, Paxton RJ. Mode of Transmission Determines the Virulence of Black Queen Cell Virus in Adult Honey Bees, Posing a Future Threat to Bees and Apiculture. Viruses. 2020; 12(5):535. https://0-doi-org.brum.beds.ac.uk/10.3390/v12050535
Chicago/Turabian StyleAl Naggar, Yahya, and Robert J. Paxton. 2020. "Mode of Transmission Determines the Virulence of Black Queen Cell Virus in Adult Honey Bees, Posing a Future Threat to Bees and Apiculture" Viruses 12, no. 5: 535. https://0-doi-org.brum.beds.ac.uk/10.3390/v12050535