Recognition of Reovirus RNAs by the Innate Immune System
Abstract
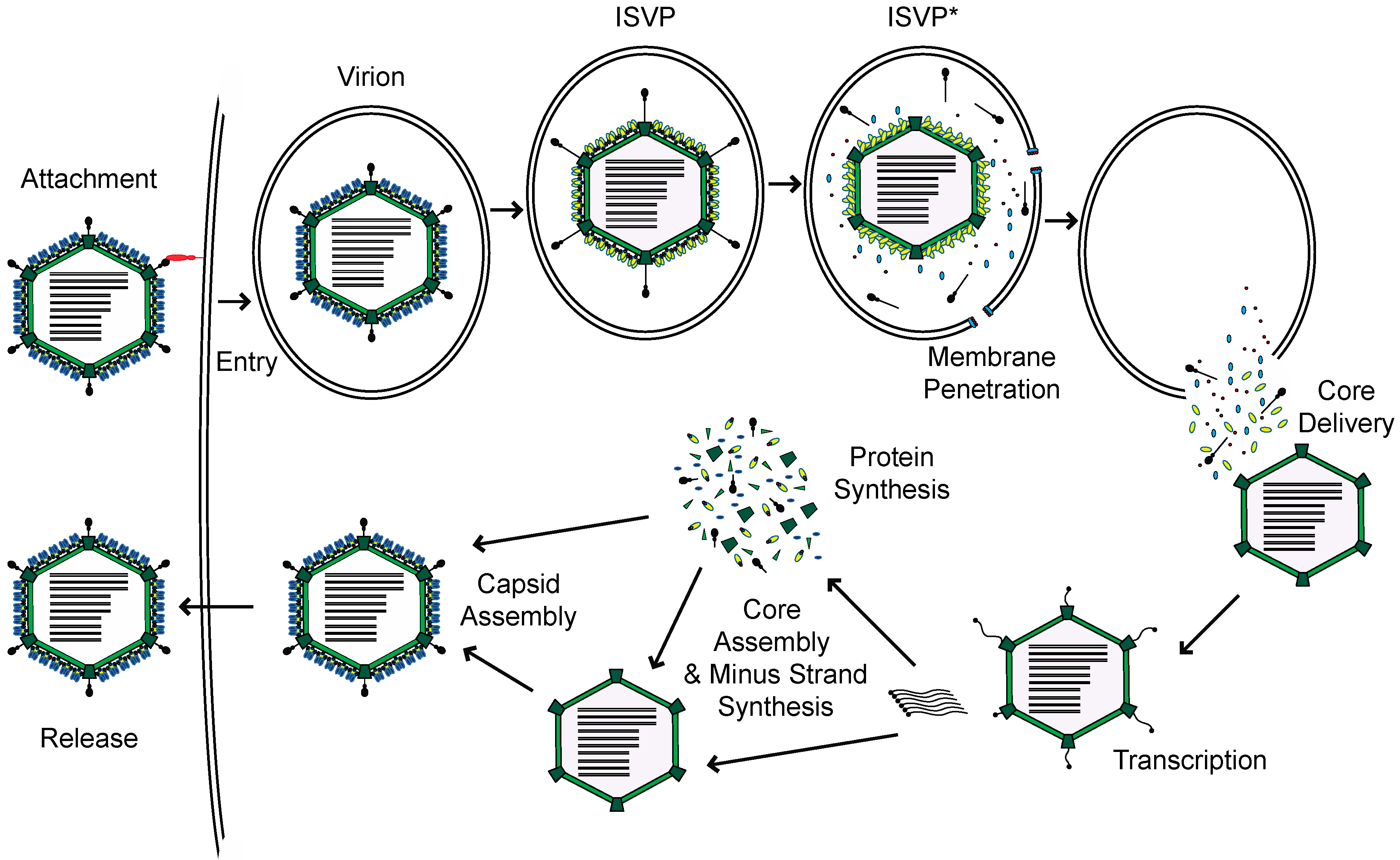
:1. Introduction
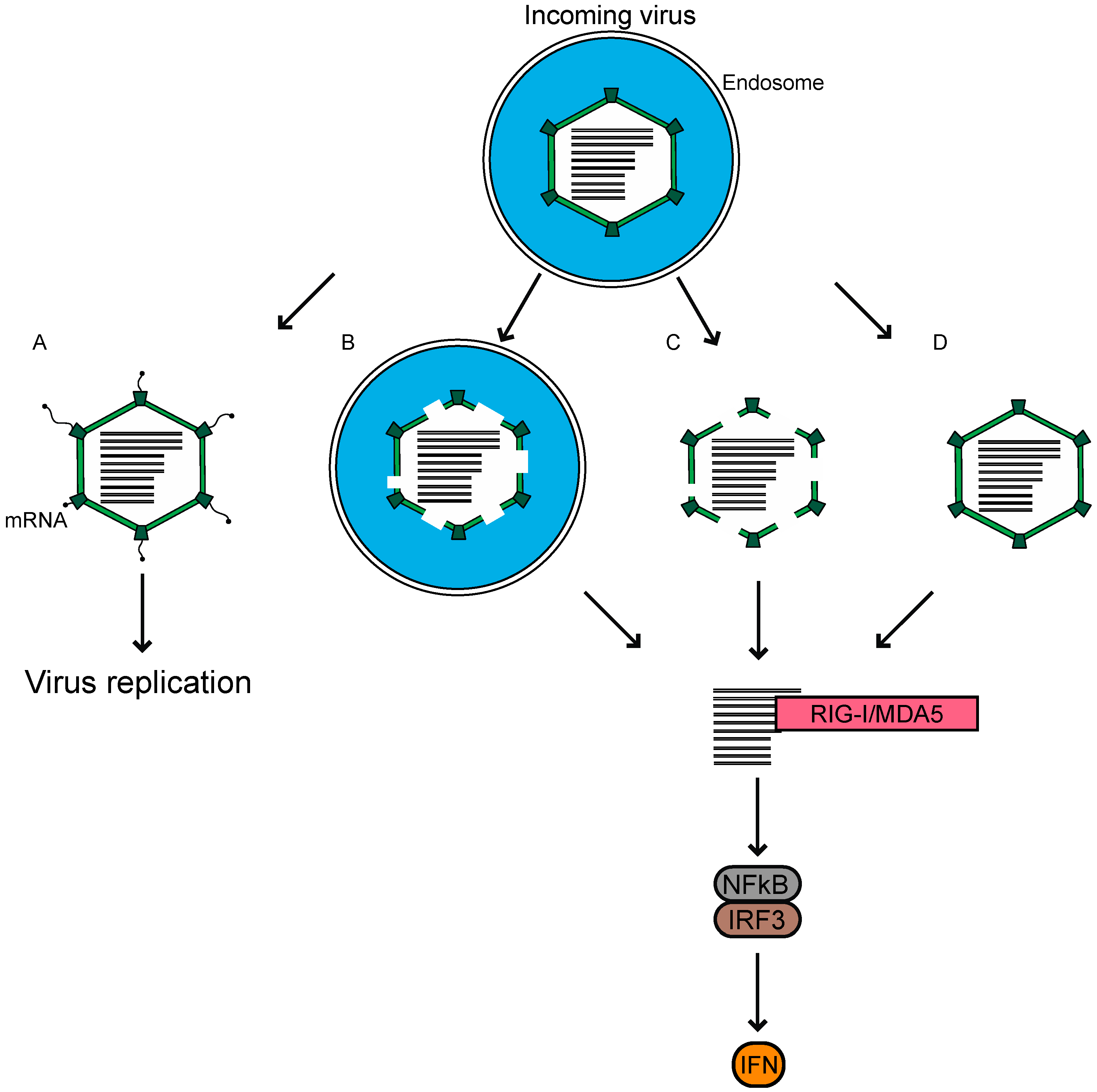
2. Sensors of Reovirus
3. Ligands
4. Sensing of Other dsRNA Viruses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Knipe, D.M.; Howley, P. Fields Virology; Wolters Kluwer Health: Philadelphia, PA, USA, 2013. [Google Scholar]
- Danthi, P.; Guglielmi, K.M.; Kirchner, E.; Mainou, B.; Stehle, T.; Dermody, T.S. From touchdown to transcription: The reovirus cell entry pathway. Curr. Top. Microbiol. Immunol. 2010, 343, 91–119. [Google Scholar] [PubMed]
- Lemay, G. Synthesis and Translation of Viral mRNA in Reovirus-Infected Cells: Progress and Remaining Questions. Viruses 2018, 10, 671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holm, G.H.; Pruijssers, A.J.; Li, L.; Danthi, P.; Sherry, B.; Dermody, T.S. Interferon Regulatory Factor 3 Attenuates Reovirus Myocarditis and Contributes to Viral Clearance. J. Virol. 2010, 84, 6900–6908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baty, C.J.; Sherry, B. Cytopathogenic effect in cardiac myocytes but not in cardiac fibroblasts is correlated with reovirus-induced acute myocarditis. J. Virol. 1993, 67, 6295–6298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goody, R.J.; Beckham, J.D.; Rubtsova, K.; Tyler, K.L. JAK-STAT signaling pathways are activated in the brain following reovirus infection. J. Neurovirology 2007, 13, 373–383. [Google Scholar] [CrossRef]
- Sherry, B.; Torres, J.; Blum, M.A. Reovirus induction of and sensitivity to beta interferon in cardiac myocyte cultures correlate with induction of myocarditis and are determined by viral core proteins. J. Virol. 1998, 72, 1314–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oie, H.K.; Leh, P.C. Reovirus Type 2: Production of and Sensitivity to Interferon in Human Amnion Cells (RA). Proc. Soc. Exp. Biol. Med. 1968, 127, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, M.T.; Lee, S.; Brown, J.J.; McAllister, N.; Urbanek, K.; Dermody, T.S.; Nice, T.J.; Virgin, H.W. Expression of Ifnlr1 on Intestinal Epithelial Cells Is Critical to the Antiviral Effects of Interferon Lambda against Norovirus and Reovirus. J. Virol. 2017, 91, e02079-16. [Google Scholar] [CrossRef] [Green Version]
- Odendall, C.; Dixit, E.; Stavru, F.; Bierne, H.; Franz, K.M.; Durbin, A.F.; Boulant, S.; Gehrke, L.; Cossart, P.; Kagan, J.C. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014, 15, 717–726. [Google Scholar] [CrossRef]
- Bender, S.; Reuter, A.; Eberle, F.; Einhorn, E.; Binder, M.; Bartenschlager, R. Activation of Type I and III Interferon Response by Mitochondrial and Peroxisomal MAVS and Inhibition by Hepatitis C Virus. PLoS Pathog. 2015, 11, e1005264. [Google Scholar] [CrossRef]
- Mahlakõiv, T.; Hernandez, P.; Gronke, K.; Diefenbach, A.; Staeheli, P. Leukocyte-Derived IFN-α/β and Epithelial IFN-λ Constitute a Compartmentalized Mucosal Defense System that Restricts Enteric Virus Infections. PLoS Pathog. 2015, 11, e1004782. [Google Scholar] [CrossRef] [PubMed]
- Pervolaraki, K.; Stanifer, M.L.; Münchau, S.; Renn, L.A.; Albrecht, D.; Kurzhals, S.; Senís, E.; Grimm, D.; Schröder-Braunstein, J.; Rabin, R.L.; et al. Type I and Type III Interferons Display Different Dependency on Mitogen-Activated Protein Kinases to Mount an Antiviral State in the Human Gut. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Zurney, J.; Howard, K.E.; Sherry, B. Basal expression levels of IFNAR and Jak-STAT components are determinants of cell-type-specific differences in cardiac antiviral responses. J. Virol. 2007, 81, 13668–13680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S.; Kawai, T.; Akira, S. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb Perspect Biol 2014, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samuel, C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef] [Green Version]
- Connolly, J.L.; Rodgers, S.E.; Clarke, P.; Ballard, D.W.; Kerr, L.D.; Tyler, K.L.; Dermody, T.S. Reovirus-induced apoptosis requires activation of transcription factor NF-kappaB. J. Virol. 2000, 74, 2981–2989. [Google Scholar] [CrossRef] [Green Version]
- Lanoie, D.; Boudreault, S.; Bisaillon, M.; Lemay, G. How Many Mammalian Reovirus Proteins are involved in the Control of the Interferon Response? Pathogens 2019, 8, 83. [Google Scholar] [CrossRef] [Green Version]
- Loo, Y.-M.; Fornek, J.; Crochet, N.; Bajwa, G.; Perwitasari, O.; Martinez-Sobrido, L.; Akira, S.; Gill, M.A.; García-Sastre, A.; Katze, M.G.; et al. Distinct RIG-I and MDA5 Signaling by RNA Viruses in Innate Immunity. J. Virol. 2008, 82, 335–345. [Google Scholar] [CrossRef] [Green Version]
- Berger, A.K.; Hiller, B.E.; Thete, D.; Snyder, A.J.; Perez, E.; Upton, J.W.; Danthi, P. Viral RNA at Two Stages of Reovirus Infection Is Required for the Induction of Necroptosis. J. Virol. 2017. [Google Scholar] [CrossRef] [Green Version]
- Holm, G.H.; Zurney, J.; Tumilasci, V.; Leveille, S.; Danthi, P.; Hiscott, J.; Sherry, B.; Dermody, T.S. Retinoic acid-inducible gene-I and interferon-beta promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J. Biol. Chem. 2007, 282, 21953–21961. [Google Scholar] [CrossRef] [Green Version]
- Goubau, D.; Schlee, M.; Deddouche, S.; Pruijssers, A.J.; Zillinger, T.; Goldeck, M.; Schuberth, C.; Van der Veen, A.G.; Fujimura, T.; Rehwinkel, J.; et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 2014, 514, 372–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuart, J.D.; Holm, G.H.; Boehme, K.W. Differential delivery of genomic dsRNA causes reovirus strain-specific differences in IRF-3 activation. J. Virol. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Yuan, B.; Lu, N.; Facchinetti, V.; Liu, Y.-J. DHX9 Pairs with IPS-1 To Sense Double-Stranded RNA in Myeloid Dendritic Cells. J. Immunol. 2011, 187, 4501–4508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Kim, T.; Bao, M.; Facchinetti, V.; Jung, S.Y.; Ghaffari, A.A.; Qin, J.; Cheng, G.; Liu, Y.J. DDX1, DDX21, and DHX36 Helicases Form a Complex with the Adaptor Molecule TRIF to Sense dsRNA in Dendritic Cells. Immunity 2011, 34, 866–878. [Google Scholar] [CrossRef] [Green Version]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Stanifer, M.L.; Rippert, A.; Kazakov, A.; Willemsen, J.; Bucher, D.; Bender, S.; Bartenschlager, R.; Binder, M.; Boulant, S. Reovirus intermediate subviral particles constitute a strategy to infect intestinal epithelial cells by exploiting TGF-β dependent pro-survival signaling. Cell. Microbiol. 2016, 18, 1831–1845. [Google Scholar] [CrossRef] [Green Version]
- Edelmann, K.H.; Richardson-Burns, S.; Alexopoulou, L.; Tyler, K.L.; Flavell, R.A.; Oldstone, M.B.A. Does Toll-like receptor 3 play a biological role in virus infections? Virology 2004, 322, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Johansson, C.; Wetzel, J.D.; He, J.P.; Mikacenic, C.; Dermody, T.S.; Kelsall, B.L. Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J. Exp. Med. 2007, 204, 1349–1358. [Google Scholar] [CrossRef] [Green Version]
- Dionne, K.R.; Galvin, J.M.; Schittone, S.A.; Clarke, P.; Tyler, K.L. Type I interferon signaling limits reoviral tropism within the brain and prevents lethal systemic infection. J. Neurovirology 2011, 17, 314–326. [Google Scholar] [CrossRef]
- Henderson, D.R.; Joklik, W.K. The mechanism of interferon induction by UV-irradiated reovirus. Virology 1978, 91, 389–406. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Mikamo-Satoh, E.; Hirai, R.; Kawai, T.; Matsushita, K.; Hiiragi, A.; Dermody, T.S.; Fujita, T.; Akira, S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008, 205, 1601–1610. [Google Scholar] [CrossRef]
- Chow, N.-L.; Shatkin, A.J. Blocked and Unblocked 5′ Termini in Reovirus Genome RNA. J. Virol. 1975, 15, 1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuichi, Y.; Muthukrishnan, S.; Shatkin, A.J. 5′-Terminal m-7G(5′)ppp(5′)G-m-p in vivo: Identification in reovirus genome RNA. Proc. Natl. Acad. Sci. USA 1975, 72, 742–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, A.K.; Shatkin, A.J. Guanosine-5′-diphosphate at the 5′ termini of reovirus RNA: Evidence for a segmented genome within the virion. J. Mol. Biol. 1971, 61, 643–653. [Google Scholar] [CrossRef]
- Miura, K.; Watanabe, K.; Sugiura, M.; Shatkin, A.J. The 5′-terminal nucleotide sequences of the double-stranded RNA of human reovirus. Proc. Natl. Acad. Sci. USA 1974, 71, 3979–3983. [Google Scholar] [CrossRef] [Green Version]
- Shatkin, A.J.; Sipe, J.D. Single-stranded, adenine-rich RNA from purified reoviruses. Proc. Natl. Acad. Sci. USA 1968, 59, 246–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, M.H.T.; Joklik, W.K. The induction of interferon by temperature-sensitive mutants of reovirus, UV-irradiated reovirus, and subviral reovirus particles. Virology 1973, 51, 191–204. [Google Scholar] [CrossRef]
- Long, W.F.; Burke, D.C. Interferon production by double-stranded RNA: A comparison of induction by reovirus to that by a synthetic double-stranded polynucleotide. J. Gen. Virol. 1971, 12, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Silversteint, S.C.; Schonberg, M.; Levin, D.H.; Acs, G. The Reovirus Replicative Cycle: Conservation of Parental RNA and Protein*. Proc. Natl. Acad. Sci. USA 1970, 67, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.T.; Zweerink, H.J. Fate of parental reovirus in infected cell. Virology 1971, 46, 544–555. [Google Scholar] [CrossRef]
- Borsa, J.; Copps, T.P.; Sargent, M.D.; Long, D.G.; Chapman, J.D. New Intermediate Subviral Particles in the In Vitro Uncoating of Reovirus Virions by Chymotrypsin. J. Virol. 1973, 11, 5244. [Google Scholar] [CrossRef] [Green Version]
- Bodkin, D.K.; Nibert, M.L.; Fields, B.N. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 1989, 63, 4676–4681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bass, D.M.; Bodkin, D.; Dambrauskas, R.; Trier, J.S.; Fields, B.N.; Wolf, J.L. Intraluminal proteolytic activation plays an important role in replication of type 1 reovirus in the intestines of neonatal mice. J. Virol. 1990, 64, 1830–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanifer, M.L.; Kischnick, C.; Rippert, A.; Albrecht, D.; Boulant, S. Reovirus inhibits interferon production by sequestering IRF-3 into viral factories. Sci. Rep. 2017, 7. [Google Scholar]
- Chandran, K.; Farsetta, D.L.; Nibert, M.L. Strategy for nonenveloped virus entry: A hydrophobic conformer of the reovirus membrane penetration protein micro 1 mediates membrane disruption. J. Virol. 2002, 76, 9920–9933. [Google Scholar] [CrossRef] [Green Version]
- Mainou, B.A.; Dermody, T.S. Transport to late endosomes is required for efficient reovirus infection. J. Virol. 2012, 86, 8346–8358. [Google Scholar] [CrossRef] [Green Version]
- Ebert, D.H.; Deussing, J.; Peters, C.; Dermody, T.S. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 2002, 277, 24609–24617. [Google Scholar] [CrossRef] [Green Version]
- Lucia-jandris, P.; Hooper, J.W.; Fields, B.N.; Borsa, J.; Morash, B.D.; Sargent, M.D.; Copps, T.P.; Lievaart, P.A.; Szekely, J.G.; Gen, J. Reovirus M2 Gene Is Associated with Chromium Release from Mouse L Cells. J. Virol. 1993, 5339–5345. [Google Scholar] [CrossRef] [Green Version]
- Hooper, J.W.; Fields, B.N. Role of the 1 Protein in Reovirus Stability and Capacity To Cause Chromium Release from Host Cells. J. Virol. 1996, 70, 459–467. [Google Scholar] [CrossRef] [Green Version]
- Boulant, S.; Stanifer, M.; Kural, C.; Cureton, D.K.; Massol, R.; Nibert, M.L.; Kirchhausen, T. Similar uptake but different trafficking and escape routes of reovirus virions and infectious subvirion particles imaged in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 2013, 24, 1196–1207. [Google Scholar] [CrossRef]
- De Boissieu, D.; Lebon, P.; Badoual, J.; Bompard, Y.; Dupont, C. Rotavirus induces alpha-interferon release in children with gastroenteritis. J. Pediatr. Gastroenterol. Nutr. 1993, 16, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Hirata, Y.; Broquet, A.H.; Menchén, L.; Kagnoff, M.F. Activation of Innate Immune Defense Mechanisms by Signaling through RIG-I/IPS-1 in Intestinal Epithelial Cells. J. Immunol. 2007, 179, 5425–5432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broquet, A.H.; Hirata, Y.; McAllister, C.S.; Kagnoff, M.F. RIG-I/MDA5/MAVS Are Required To Signal a Protective IFN Response in Rotavirus-Infected Intestinal Epithelium. J. Immunol. 2011, 186, 1618–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, A.; Pruijssers, A.J.; Dermody, T.S.; García-Sastre, A.; Greenberg, H.B. The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS-1, RIG-I, MDA-5, and IRF-3. J. Virol. 2011, 85, 3717–3732. [Google Scholar] [CrossRef] [Green Version]
- Uzri, D.; Greenberg, H.B. Characterization of Rotavirus RNAs That Activate Innate Immune Signaling through the RIG-I-Like Receptors. PLoS ONE 2013, 8, e69825. [Google Scholar] [CrossRef] [Green Version]
- Gallegos, C.O.; Patton, J.T. Characterization of rotavirus replication intermediates: A model for the assembly of single-shelled particles. Virology 1989, 172, 616–627. [Google Scholar] [CrossRef]
- Patton, J.T.; Gallegos, C.O. Rotavirus RNA replication: Single-stranded RNA extends from the replicase particle. J. Gen. Virol. 1990, 71, 1087–1094. [Google Scholar] [CrossRef]
- Rojas, M.; Arias, C.F.; López, S. Protein kinase R is responsible for the phosphorylation of eIF2alpha in rotavirus infection. J. Virol. 2010, 84, 10457–10466. [Google Scholar] [CrossRef] [Green Version]
- McKimm-Breschkin, J.L.; Holmes, I.H. Conditions required for induction of interferon by rotaviruses and for their sensitivity to its action. Infect. Immun. 1982, 36, 857–863. [Google Scholar] [CrossRef] [Green Version]
- Rollo, E.E.; Kumar, K.P.; Reich, N.C.; Cohen, J.; Angel, J.; Greenberg, H.B.; Sheth, R.; Anderson, J.; Oh, B.; Hempson, S.J.; et al. The Epithelial Cell Response to Rotavirus Infection. J. Immunol. 1999, 163, 4442–4452. [Google Scholar]
- Deal, E.M.; Jaimes, M.C.; Crawford, S.E.; Estes, M.K.; Greenberg, H.B. Rotavirus structural proteins and dsRNA are required for the human primary plasmacytoid dendritic cell IFNalpha response. PLoS Pathog. 2010, 6, e1000931. [Google Scholar] [CrossRef] [Green Version]
- Frias, A.H.; Vijay-Kumar, M.; Gentsch, J.R.; Crawford, S.E.; Carvalho, F.A.; Estes, M.K.; Gewirtz, A.T. Intestinal epithelia activate anti-viral signaling via intracellular sensing of rotavirus structural components. Mucosal Immunol. 2010, 3, 622–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, M.M.; Sen, A.; Greenberg, H.B.; Patton, J.T. The Battle between Rotavirus and Its Host for Control of the Interferon Signaling Pathway. PloS Pathog. 2013, 9, e1003064. [Google Scholar] [CrossRef]
- Benavente, J.; Martínez-Costas, J. Avian reovirus: Structure and biology. Virus Res. 2007, 123, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.N.; Eidson, C.S.; Brown, J.; Kleven, S.H. Studies on interferon induction and interferon sensitivity of avian reoviruses. Avian Dis. 1983, 27, 927–936. [Google Scholar] [CrossRef]
- Lostalé-Seijo, I.; Martínez-Costas, J.; Benavente, J. Response of Three Different Viruses to Interferon Priming and Dithiothreitol Treatment of Avian Cells. J. Virol. 2016, 90, 8328–8340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lostalé-Seijo, I.; Martínez-Costas, J.; Benavente, J. Interferon induction by avian reovirus. Virology 2016, 487, 104–111. [Google Scholar] [CrossRef] [Green Version]
- Neerukonda, S.N.; Katneni, U. Avian Pattern Recognition Receptor Sensing and Signaling. Vet. Sci. 2020, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Huismans, H. Bluetongue virus-induced interferon synthesis. Onderstepoort J. Vet. Res. 1969, 36, 181–185. [Google Scholar]
- MacLachlan, N.J.; Schore, C.E.; Osburn, B.I. Antiviral responses of bluetongue virus-inoculated bovine fetuses and their dams. Am. J. Vet. Res. 1984, 45, 1469–1473. [Google Scholar]
- Chauveau, E.; Doceul, V.; Lara, E.; Adam, M.; Breard, E.; Sailleau, C.; Viarouge, C.; Desprat, A.; Meyer, G.; Schwartz-Cornil, I.; et al. Sensing and Control of Bluetongue Virus Infection in Epithelial Cells via RIG-I and MDA5 Helicases. J. Virol. 2012, 86, 11789–11799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruscanu, S.; Pascale, F.; Bourge, M.; Hemati, B.; Elhmouzi-Younes, J.; Urien, C.; Bonneau, M.; Takamatsu, H.; Hope, J.; Mertens, P.; et al. The double-stranded RNA bluetongue virus induces type I interferon in plasmacytoid dendritic cells via a MYD88-dependent TLR7/8-independent signaling pathway. J. Virol. 2012, 86, 5817–5828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jameson, P.; Schoenherr, C.K.; Grossberg, S.E. Bluetongue virus, an exceptionally potent interferon inducer in mice. Infect. Immun. 1978, 20, 321–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitour, D.; Doceul, V.; Ruscanu, S.; Chauveau, E.; Schwartz-Cornil, I.; Zientara, S. Induction and control of the type I interferon pathway by Bluetongue virus. Virus Res. 2014, 182, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Dales, S.; Omatos, P.J.; Hsu, K.C. The uptake and development of reovirus in strain L cells followed with labeled viral ribonucleic acid and ferritin-antibody conjugates. Virology 1965, 25, 193–211. [Google Scholar] [CrossRef]
- Silverstein, S.C.; Dales, S. The penetration of reovirus RNA and initiation of its genetic function in L-strain fibroblasts. J. Cell Biol. 1968, 36, 197–230. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abad, A.T.; Danthi, P. Recognition of Reovirus RNAs by the Innate Immune System. Viruses 2020, 12, 667. https://0-doi-org.brum.beds.ac.uk/10.3390/v12060667
Abad AT, Danthi P. Recognition of Reovirus RNAs by the Innate Immune System. Viruses. 2020; 12(6):667. https://0-doi-org.brum.beds.ac.uk/10.3390/v12060667
Chicago/Turabian StyleAbad, Andrew T., and Pranav Danthi. 2020. "Recognition of Reovirus RNAs by the Innate Immune System" Viruses 12, no. 6: 667. https://0-doi-org.brum.beds.ac.uk/10.3390/v12060667




