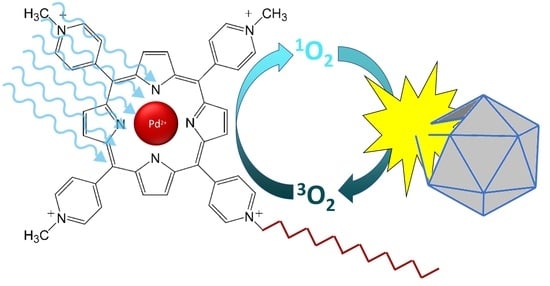
A Comparison of Porphyrin Photosensitizers in Photodynamic Inactivation of RNA and DNA Bacteriophages
Abstract
:1. Introduction
2. Methods
2.1. Porphyrins and Metalloporphyrin Synthesis
2.2. Bacteriophage Propagation
2.3. Absorbance Spectra
2.4. Photodynamic Inactivation
2.5. Data Analysis
3. Results and Discussion
3.1. Confirmation of Free and Bound Porphyrin in Test Conditions
3.2. Relative Resistance of Bacteriophages to PDI
3.3. Comparative Efficacy of Different Porphyrins for Virus Inactivation
3.4. Efficacy of Bacteriophage Inactivation Compared to Similar Techniques
4. Conclusions
- Both inclusion of a hydrophobic C14 chain and/or a central palladium ion in the porphyrin structure significantly improves bacteriophage inactivation using low-dose 405 nm illumination.
- Inactivation rates differed by two orders of magnitude among bacteriophages. Susceptibility to PDI is not entirely predictable by genome type; however, ssRNA bacteriophages are particularly susceptible and are therefore poor surrogates for human viruses of interest. In this study, bacteriophage ΦX174 was the most resistant, with inactivation rates comparable to human viruses of interest in the literature. Therefore, ΦX174 should be considered as a virus surrogate for PDI research.
- Inactivation rates using the novel PdT4, PdC14, and C14 porphyrins and blue-light LED were favorable in comparison with previous reports in the literature.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization Viruses. In Water Recreation and Disease; IWA Publishing: London, UK, 2005; pp. 191–229. ISBN 1843390663.
- Pang, X.L.; Lee, B.E.; Pabbaraju, K.; Gabos, S.; Craik, S.; Payment, P.; Neumann, N. Pre-analytical and analytical procedures for the detection of enteric viruses and enterovirus in water samples. J. Virol. Methods 2012, 184, 77–83. [Google Scholar] [CrossRef]
- Abbaszadegan, M.; Lechevallier, M.; Gerba, C.P. Occurrence of viruses in US groundwaters. J. Am. Water Works Assoc. 2003, 95, 107–120. [Google Scholar] [CrossRef]
- Hall, A.J. Noroviruses: The perfect human pathogens? J. Infect. Dis. 2012, 205, 1622–1624. [Google Scholar] [CrossRef] [Green Version]
- Boyd, E.F. Chapter 4—Bacteriophage-encoded bacterial virulence factors and phage–pathogenicity island interactions. In Bacteriophages, Part A; Łobocka, M., Szybalski, W., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 82, pp. 91–118. ISBN 0065-3527. [Google Scholar]
- Lood, R.; Ertürk, G.; Mattiasson, B. Revisiting antibiotic resistance spreading in wastewater treatment plants—Bacteriophages as a much neglected potential transmission vehicle. Front. Microbiol. 2017, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Majiya, H.; Chowdhury, K.F.; Stonehouse, N.J.; Millner, P. TMPyP functionalised chitosan membrane for efficient sunlight driven water disinfection. J. Water Process Eng. 2019, 30, 100475. [Google Scholar] [CrossRef]
- Costa, L.; Alves, E.; Carvalho, C.M.B.; Tome, J.P.C.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Tome, A.C.; Cavaleiro, J.A.S.; Cunha, A.; Almeida, A. Sewage bacteriophage photoinactivation by cationic porphyrins: A study of charge effect. Photochem. Photobiol. Sci. 2008, 7, 415–422. [Google Scholar] [CrossRef]
- Costa, L.; Carvalho, C.M.B.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Tome, J.P.C.; Tome, A.C.; Cavaleiro, J.A.S.; Cunha, A.; Almeida, A. Sewage bacteriophage inactivation by cationic porphyrins: Influence of light parameters. Photochem. Photobiol. Sci. 2010, 9, 1126–1133. [Google Scholar] [CrossRef] [PubMed]
- Alves, E.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Cunha, Â.; Nadais, H.; Almeida, A. Potential applications of porphyrins in photodynamic inactivation beyond the medical scope. J. Photochem. Photobiol. C Photochem. Rev. 2015, 22, 34–57. [Google Scholar] [CrossRef] [Green Version]
- Peddinti, B.S.T.; Scholle, F.; Ghiladi, R.A.; Spontak, R.J. Photodynamic polymers as comprehensive anti-Infective materials: Staying ahead of a growing global threat. ACS Appl. Mater. Interfaces 2018, 10, 25955–25959. [Google Scholar] [CrossRef]
- Zupán, K.; Egyeki, M.; Tóth, K.; Fekete, A.; Herényi, L.; Módos, K.; Csík, G. Comparison of the efficiency and the specificity of DNA-bound and free cationic porphyrin in photodynamic virus inactivation. J. Photochem. Photobiol. B Biol. 2008, 90, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Rywkin, S.; Ben-Hur, E.; Malik, Z.; Prince, A.M.; Li, Y.-S.; Kenney, M.E.; Oleinick, N.L.; Horowitz, B. New phthalocyanines for photodynamic virus inactivation in red blood cell concentrates. Photochem. Photobiol. 1994, 60, 165–190. [Google Scholar] [CrossRef]
- Casteel, M.J.; Jayaraj, K.; Gold, A.; Ball, L.M.; Sobsey, M.D. Photoinactivation of hepatitis A virus by synthetic porphyrins T. Photochem. Photobiol. 2004, 80, 294–300. [Google Scholar] [CrossRef]
- Dolphin, D. The Porphyrins V5: Physical Chemistry, Part C; Academic Press Inc.: New York, NY, USA, 2012; ISBN 9780323145725. [Google Scholar]
- Pandey, R.K.; Dougherty, T.J.; Kessel, D. Handbook of Photodynamic Therapy: Updates on Recent Applications of Porphyrin-Based Compounds; World Scientific Publishing Company: Singapore, 2016; ISBN 9789814719667. [Google Scholar]
- Majiya, H.; Adeyemi, O.O.; Stonehouse, N.J.; Millner, P. Photodynamic inactivation of bacteriophage MS2: The A-protein is the target of virus inactivation. J. Photochem. Photobiol. B Biol. 2018, 178, 404–411. [Google Scholar] [CrossRef] [Green Version]
- Leviskas, B.; Valyi-Nagy, T.; Munirathinam, G.; Bork, M.; Valyi-Nagy, K.; Skwor, T. Metalloporphyrin Pd(T4) exhibits oncolytic activity and cumulative effects with 5-ALA photodynamic treatment against C918 cells. Int. J. Mol. Sci. 2020, 21, 669. [Google Scholar] [CrossRef] [Green Version]
- Costa, L.; Esteves, A.C.; Correia, A.; Moreirinha, C.; Delgadillo, I.; Cunha, Â.; Neves, M.G.P.S.; Faustino, M.A.F.; Almeida, A. SDS-PAGE and IR spectroscopy to evaluate modifications in the viral protein profile induced by a cationic porphyrinic photosensitizer. J. Virol. Methods 2014, 209, 103–109. [Google Scholar] [CrossRef]
- Shnipper, L.E.; Lewin, A.A.; Schwartz, M.; Crumpacker, C.S. Mechanisms of photodynamic inactivation of herpes simplex viruses. J. Clin. Investig. 1980, 65, 432–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, L.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Cunha, Â.; Almeida, A. Photodynamic inactivation of mammalian viruses and bacteriophages. Viruses 2012, 4, 1034–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, M.; Lee, J.; Mackeyev, Y.; Wilson, L.J.; Alvarez, P.J.J.; Hughes, J.B.; Kim, J.-H. Visible light sensitized inactivation of MS-2 bacteriophage by a cationic amine-functionalized C60 derivative. Environ. Sci. Technol. 2010, 44, 6685–6691. [Google Scholar] [CrossRef]
- Costa, L.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Tomé, A.C.; Cavaleiro, J.A.S.; Cunha, Â.; Faustino, M.A.F.; Almeida, A. Susceptibility of non-enveloped DNA- and RNA-type viruses to photodynamic inactivation. Photochem. Photobiol. Sci. 2012, 11, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Moor, A.C.E.; Wagenaars-van Gompell, A.E.; Brand, A.; Dubbelmanl, T.M.A.R.; VanSteveninck, J. Primary targets for photoinactivation of vesicular stomatitis virus by AlPcS4 or Pc4 and red light. Photochem. Photobiol. 1997, 65, 465–470. [Google Scholar] [CrossRef]
- Majiya, H.; Adeyemi, O.O.; Herod, M.; Stonehouse, N.J.; Millner, P. Photodynamic inactivation of non-enveloped RNA viruses. J. Photochem. Photobiol. B Biol. 2018, 189, 87–94. [Google Scholar] [CrossRef]
- Remichkova, M.; Mukova, L.; Nikolaeva-glomb, L.; Nikolova, N.; Doumanova, L.; Mantareva, V.; Angelov, I.; Kussovski, V.; Galabov, A.S. Virus inactivation under the photodynamic effect of phthalocyanine zinc(II) complexes. Z. Naturforsch 2017, 72, 123–128. [Google Scholar] [CrossRef]
- Monjo, A.L.A.; Pringle, E.S.; Thornbury, M.; Duguay, B.A.; Monro, S.M.A.; Hetu, M.; Knight, D.; Cameron, C.G.; McFarland, S.A.; McCormick, C. Photodynamic inactivation of herpes simplex viruses. Viruses 2018, 10, 532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korneev, D.; Kurskaya, O.; Sharshov, K.; Eastwood, J.; Strakhovskaya, M. Ultrastructural aspects of photodynamic inactivation of highly pathogenic avian H5N8 influenza virus. Viruses 2019, 11, 955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skwor, T.A.; Klemm, S.; Zhang, H.; Schardt, B.; Blaszczyk, S.; Bork, M.A. Photodynamic inactivation of methicillin-resistant Staphylococcus aureus and Escherichia coli: A metalloporphyrin comparison. J. Photochem. Photobiol. B Biol. 2016, 165, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Bork, M.A.; Gianopoulos, C.G.; Zhang, H.; Fanwick, P.E.; Choi, J.H.; McMillin, D.R. Accessibility and external versus intercalative binding to DNA as assessed by oxygen-induced quenching of the palladium(II)-containing cationic porphyrins Pd(T4) and Pd(t D4). Biochemistry 2014, 53, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Baier, J.; Fuß, T.; Pöllmann, C.; Wiesmann, C.; Pindl, K.; Engl, R.; Baumer, D.; Maier, M.; Landthaler, M.; Bäumler, W. Theoretical and experimental analysis of the luminescence signal of singlet oxygen for different photosensitizers. J. Photochem. Photobiol. B Biol. 2007, 87, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Mayer, B.K.; Ryu, H.; Abbaszadegan, M. Treatability of U.S. Environmental Protection Agency contaminant candidate list viruses: Removal of coxsackievirus and echovirus using enhanced coagulation. Environ. Sci. Technol. 2008, 42, 6890–6896. [Google Scholar] [CrossRef] [Green Version]
- Mayer, B.K.; Yang, Y.; Gerrity, D.W.; Abbaszadegan, M. The impact of capsid proteins on virus removal and inactivation during water treatment processes. Microbiol. Insights 2015, 8, 15–28. [Google Scholar] [CrossRef]
- Armanious, A.; Aeppli, M.; Jacak, R.; Refardt, D.; Sigstam, T.; Kohn, T.; Sander, M. Viruses at solid-water interfaces: A systematic assessment of interactions driving adsorption. Environ. Sci. Technol. 2016, 50, 732–743. [Google Scholar] [CrossRef]
- Shen, C.; Phanikumar, M.S.; Fong, T.-T.; Aslam, I.; Mcelmurry, S.P.; Molloy, S.L.; Rose, J.B. Evaluating bacteriophage P22 as a tracer in a complex surface water system: The Grand River, Michigan. Environ. Sci. Technol. 2008, 42, 2426–2431. [Google Scholar] [CrossRef] [PubMed]
- De Fidalgo Cortalezzi, M.M.; Gallardo, M.V.; Yrazu, F.; Gentile, G.J.; Opezzo, O.; Pizarro, R.; Poma, H.R.; Rajal, V.B. Virus removal by iron oxide ceramic membranes. J. Environ. Chem. Eng. 2014, 2, 1831–1840. [Google Scholar] [CrossRef]
- Michen, B.; Graule, T. Isoelectric points of viruses. J. Appl. Microbiol. 2010, 109, 388–397. [Google Scholar] [CrossRef] [Green Version]
- Csík, G.; Egyeki, M.; Herényi, L.; Majer, Z.; Tóth, K. Role of structure-proteins in the porphyrin-DNA interaction. J. Photochem. Photobiol. B Biol. 2009, 96, 207–215. [Google Scholar] [CrossRef]
- Beck, N.K.; Callahan, K.; Nappier, S.P.; Kim, H.; Sobsey, M.D.; Meschke, J.S. Development of a spot-titer culture assay for quantifying bacteria and viral indicators. J. Rapid Methods Autom. Microbiol. 2009, 17, 455–464. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An {R} Companion to Applied Regression, 2nd ed.; Sage: Thousand Oaks, CA, USA, 2011. [Google Scholar]
- Lenth, R.V. Least-Squares Means: The R Package lsmeans. J. Stat. Softw. 2016, 69, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Hotze, E.M.; Badireddy, A.R.; Chellam, S.; Wiesner, M.R. Mechanisms of bacteriophage inactivation via singlet oxygen generation in UV illuminated fullerol suspensions. Environ. Sci. Technol. 2009, 43, 6639–6645. [Google Scholar] [CrossRef]
- Heffron, J.; Mayer, B.K. Improved virus isoelectric point estimation by exclusion of known and predicted genome-binding regions. Appl. Environ. Microbiol. 2020, 86, e01674-20. [Google Scholar] [CrossRef]
- Heffron, J.; Mayer, B.K. Virus isoelectric point estimation: Theories and methods. Appl. Environ. Microbiol. 2021, 87, e02319-20. [Google Scholar] [CrossRef]
- Schneider, J.E.; Tabatabaie, T.; Maidt, L.; Smith, R.H.; Nguyen, X.; Pye, Q.; Floyd, R.A. Potential mechanisms of photodynamic inactivation of virus by methylene blue I. RNA-protein crosslinks and other oxidative lesions in Qβ bacteriophage. Photochem. Photobiol. 1998, 67, 350–357. [Google Scholar] [CrossRef]
- Cerritelli, M.E.; Cheng, N.; Rosenberg, A.H.; Mcpherson, C.E.; Booy, F.P.; Steven, A.C. Encapsidated conformation of bacteriophage T7 DNA. Cell 1997, 91, 271–280. [Google Scholar] [CrossRef] [Green Version]
- Obrien, B.J.M.; Gaffney, D.K.; Wang, T.P.; Sieber, F. Merocyanine 540-sensitized photoinactivation of enveloped viruses in blood products: Site and mechanism of phototoxicity. Blood 1992, 80, 277–285. [Google Scholar] [CrossRef] [Green Version]
- Käsermann, F.; Kempf, C. Photodynamic inactivation of enveloped viruses by buckminsterfullerene. Antiviral Res. 1997, 34, 65–70. [Google Scholar] [CrossRef]
- Silva, E.M.P.; Giuntini, F.; Faustino, M.A.F.; Tome, P.C.; Silva, A.M.S.; Neves, M.G.P.M.S.; Tome, A.C.; Ferrer-Correia, J.; Santana-marques, M.G.; Caeiro, M.F.; et al. Synthesis of cationic b-vinyl substituted meso-tetraphenylporphyrins and their in vitro activity against herpes simplex virus type 1. Bioorg. Med. Chem. 2005, 15, 3333–3337. [Google Scholar] [CrossRef] [PubMed]
- Nikolaeva-Glomb, L.; Mukova, L.; Nikolova, N.; Kussovski, V.; Doumanova, L.; Mantareva, V.; Angelov, I.; Wohrle, D.; Galabov, A.S. Photodynamic effect of some phthalocyanines on enveloped and naked viruses. Acta Virol. 2017, 61, 341–346. [Google Scholar] [CrossRef] [Green Version]
- Heffron, J.; McDermid, B.; Maher, E.; McNamara, P.; Mayer, B.K. Mechanisms of virus mitigation and suitability of bacteriophages as surrogates in drinking water treatment by iron electrocoagulation. Water Res. 2019, 163, 114877. [Google Scholar] [CrossRef] [PubMed]
- Kreißel, K.; Bösl, M.; Hügler, M.; Lipp, P.; Franzreb, M.; Hambsch, B. Inactivation of F-specific bacteriophages during flocculation with polyaluminum chloride—A mechanistic study. Water Res. 2014, 51, 144–151. [Google Scholar] [CrossRef]
- Gerba, C.P.; Betancourt, W.Q. Viral Aggregation: Impact on Virus Behavior in the Environment. Environ. Sci. Technol. 2017, 51, 7318–7325. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Li, H.; Yang, M.; Wu, F. Palladium porphyrin complexes for photodynamic cancer therapy: Effect of porphyrin units and metal. Photochem. Photobiol. Sci. 2020, 19, 905–912. [Google Scholar] [CrossRef]
- Rapozzi, V.; Zorzet, S.; Zacchigna, M.; Della Pietra, E.; Cogoi, S.; Xodo, L.E. Anticancer activity of cationic porphyrins in melanoma tumour-bearing mice and mechanistic in vitro studies. Mol. Cancer 2014, 13, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Wagner, S.J.; Skripchenkol, A.; Robinenel, D.; Foley, J.W.; Cincotta, L. Factors affecting virus photoinactivation by a series of phenothiazine dyes. Photochemistry 1998, 67, 343–349. [Google Scholar] [CrossRef]
- Davies, M.J. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 2003, 305, 761–770. [Google Scholar] [CrossRef]
- Wilkinson, F.; Helman, W.P.; Ross, A.B. Rate constants for the decay and reactions of the lowest electronically excited singlet state of molecular oxygen in solution. An expanded and revised compilation. J. Phys. Chem. Ref. Data 1995, 24, 663–1021. [Google Scholar] [CrossRef] [Green Version]
- Pasternack, R.F.; Caccam, M.; Keogh, B.; Stephenson, T.A.; Williams, A.P.; Gibbs, E.J. Long-range fluorescence quenching of ethidium ion by cationic porphyrins in the presence of DNA. J. Am. Chem. Soc. 1991, 113, 6835–6840. [Google Scholar] [CrossRef]
- Kano, K.; Minamizono, H.; Kitae, T.; Negi, S. Self-aggregation of cationic porphyrins in water. Can π-π stacking interaction overcome electrostatic repulsive force? J. Phys. Chem. A 1997, 101, 6118–6124. [Google Scholar] [CrossRef]
- Han, H.; Langley, D.R.; Rangan, A.; Hurley, L.H. Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc. 2001, 123, 8902–8913. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Song, D.; Qin, T.; Yang, C.; Yu, Z.; Li, X.; Liu, K.; Su, H. Interaction between G-quadruplex and zinc cationic porphyrin: The Role of the Axial Water. Sci. Rep. 2017, 7, 2–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.R.; Min, Y.Q.; Wang, J.Q.; Liu, C.X.; Fu, B.S.; Wu, F.; Wu, L.Y.; Qiao, Z.X.; Song, Y.Y.; Xu, G.H.; et al. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti–hepatitis C target. Sci. Adv. 2016, 2. [Google Scholar] [CrossRef] [Green Version]
- Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N.P.; Clancy, J.L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10, 358–364. [Google Scholar] [CrossRef]
- Horsburgh, B.C.; Kollmus, H.; Hauser, H.; Coen, D.M. Translational recoding induced by G-rich mRNA sequences that form unusual structures. Cell 1996, 86, 949–959. [Google Scholar] [CrossRef] [Green Version]
- Schagen, F.H.E.; Moor, A.C.E.; Cheong, S.C.; Cramer, S.J.; Van Ormondt, H.; Van Der Eb, A.J.; Dubbelman, T.M.A.R.; Hoeben, R.C. Photodynamic treatment of adenoviral vectors with visible light: An easy and convenient method for viral inactivation. Gene Ther. 1999, 6, 873–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grubaugh, N.D.; Ladner, J.T.; Lemey, P.; Pybus, O.G.; Rambaut, A.; Holmes, E.C.; Andersen, K.G. Tracking virus outbreaks in the twenty-first century. Nat. Microbiol. 2019, 4, 10–19. [Google Scholar] [CrossRef] [PubMed]



| Bacteriophage | Host Bacteria | Baltimore Classification | Diameter (nm) | Isoelectric Point |
|---|---|---|---|---|
| fr (ATCC 15767-B1) | E. coli (ATCC 19853) | IV ((+)ssRNA) | 19–23 | 3.5 [35] |
| P22 (ATCC 19585-B1) | Salmonella enterica subsp. typhimurium LT2 (ATCC 19585) | I (dsDNA) | 52–60 [36] | 3.4 [37] |
| ΦX174 (ATCC 13706-B1) | E. coli (ATCC 13706) | II (ssDNA) | 23–27 | 6.0–7.0 [38] |
| Source | Light | Intensity (mW cm−2) | Compound * | Conc. (µM) | Virus | Genome | Pseudo-First Order Rate Constant (log10 s−1) | Normalized Rate Constant (log10 L cm2 µmol−1 mJ−1) |
|---|---|---|---|---|---|---|---|---|
| Enveloped mammalian viruses | ||||||||
| Obrien 1992 [48] | White (fluorescent) | 7 | Merocyanine 540 | 26 | Herpes simplex 1 virus | dsDNA | 2 × 10−2 | 9 × 10−5 |
| Käsermann 1997 [49] | White (mercury) | 29 | Fullerene | 1400 | Vesicular stomatitis virus | ssRNA | 5 × 10−4 | 1 × 10−8 |
| Semliki Forest virus | ssRNA | 5 × 10−4 | 1 × 10−8 | |||||
| Moor 1997 [24] | Red (halogen) | 46 | AlPcS4 | 1 | Vesicular Stomatitis virus | ssRNA | 1 × 10−2 | 3 × 10−4 |
| Pc4 | 0.005 | 7 × 10−3 | 3 × 10−2 | |||||
| Silva 2005 [50] | White | 50 | Pyridylvinyl-substituted tetraphenol porphryin | 0.5 | Herpes simplex 1 virus | dsDNA | 2 × 10−3 | 9 × 10−5 |
| Peddinti 2008 [11] | White (400-700 nm) | 80 | ZnTMPyP | 1% wt film | Vesicular Stomatitis virus | ssRNA | 2 × 10−3 | NA |
| Nikolaeva-Glomb 2017 [51] | Red (laser, 635nm) | 100 | Hematoporphyrin | 20 | Influenza virus A | ssRNA | 6 × 10−4 | 3 × 10−7 |
| Bovine viral diarrhea virus | ssRNA | 2 × 10−3 | 8 × 10−7 | |||||
| GaPc1 | 1 × 10−3 | 6 × 10−7 | ||||||
| GaPc2 | 2 × 10−3 | 1 × 10−6 | ||||||
| InPc1 | 2 × 10−3 | 8 × 10−7 | ||||||
| Remichkova 2017 [26] | Red (laser, 635nm) | 100 | ZnPcMe | 0.58 | Bovine viral diarrhea virus | ssRNA | 7 × 10−3 | 1 × 10−4 |
| Herpes simplex 1 virus | dsDNA | 1 × 10−2 | 2 × 10−4 | |||||
| Vaccinia virus | dsDNA | 7 × 10−3 | 1 × 10−4 | |||||
| Newcastle disease virus | ssRNA | 0 | 0 | |||||
| ZnPcS | Bovine viral diarrhea virus | ssRNA | 2 × 10−2 | 3 × 10−4 | ||||
| Herpes simplex 1 virus | dsDNA | 1 × 10−2 | 2 × 10−4 | |||||
| Vaccinia virus | dsDNA | 7 × 10−3 | 1 × 10−4 | |||||
| Newcastle disease virus | ssRNA | 3 × 10−3 | 6 × 10−5 | |||||
| Nonenveloped mammalian viruses | ||||||||
| chagen 1999 [52] | White (halogen) | 106 | Methylene blue | 1.3 | Recombinant adenovirus (E1 deficient) | dsDNA | 2 × 10−2 | 1 × 10−4 |
| Rose bengal | 10 | 7 × 10−3 | 6 × 10−6 | |||||
| Uroporphyrin | 20 | 5 × 10−3 | 2 × 10−6 | |||||
| AlPcS4 | 10 | 3 × 10−3 | 3 × 10−6 | |||||
| Peddinti 2008 [11] | White | 80 | ZnTMPyP4+ | 1% wt film | Human adenovirus 5 | dsDNA | 1 × 10−3 | NA |
| Nikolaeva-Glomb 2017 [51] | Red (laser, 635nm) | 100 | GaPc1 | 20 | Human adenovirus 5 | dsDNA | 2 × 10−3 | 8 × 10−7 |
| Poliovirus 1 | ssRNA | 1 × 10−3 | 6 × 10−7 | |||||
| GaPc2 | Human adenovirus 5 | dsDNA | 2 × 10−3 | 1 × 10−6 | ||||
| Poliovirus 1 | ssRNA | 6 × 10−4 | 3 × 10−7 | |||||
| Hematoporphyrin | Human adenovirus 5 | dsDNA | 1 × 10−3 | 6 × 10−7 | ||||
| Poliovirus 1 | ssRNA | 6 × 10−4 | 3 × 10−7 | |||||
| InPc1 | Human adenovirus 5 | dsDNA | 1 × 10−3 | 6 × 10−7 | ||||
| Poliovirus 1 | ssRNA | 6 × 10−4 | 3 × 10−7 | |||||
| Remichkova 2017 [26] | Red (laser, 635nm) | 100 | ZnPcMe | 0.58 | Coxsackievirus B1 | ssRNA | 0 | 0 |
| Human adenovirus 5 | dsDNA | 8 × 10−4 | 1 × 10−5 | |||||
| Majiya 2018 [25] | White (fluorescent) | 32 | TMPyP | 5 | Murine norovirus-1 | ssRNA | 2 × 10−3 | 1 × 10−5 |
| 10 | Bovine enterovirus-2 | ssRNA ssRNA | 2 × 10−3 | 5 × 10−6 | ||||
| 5 | 3 × 10−4 | 2 × 10−6 | ||||||
| 10 | ssRNA | 8 × 10−4 | 3 × 10−6 | |||||
| Nonenveloped bacteriophages | ||||||||
| Cho 2010 [22] | White (fluorescent) | 0.2 | Amine-functionalized fullerol | 20 | MS2 phage | ssRNA | 1 × 10−3 | 3 × 10−4 |
| Sunlight | 0.19 | 10 | 1 × 10−3 | 6 × 10−4 | ||||
| Costa 2012 [23] | White (fluorescent) | 40 | Tri-Py+-Me-PF | 5 | T4-like phage | dsDNA | 7 × 10−4 | 4 × 10−6 |
| 5 | Aeromonas phage | dsDNA | 2 × 10−4 | 9 × 10−7 | ||||
| 5 | Vibrio phage | dsDNA | 4 × 10−4 | 2 × 10−6 | ||||
| 5 | Pseudomonas phage | dsDNA | 4 × 10−4 | 2 × 10−6 | ||||
| 0.5 | MS2 phage | ssRNA | 4 × 10−3 | 2 × 10−4 | ||||
| 0.5 | Qbeta phage | ssRNA | 4 × 10−3 | 2 × 10−4 | ||||
| 0.5 | LAIST_PG002 phage | ssRNA | 7 × 10−3 | 3 × 10−4 | ||||
| Majiya 2018 [25] | White (fluorescent) | 32 | TMPyP | 0.5 | Qbeta phage | ssRNA | 2 × 10−2 | 1 × 10−3 |
| Majiya 2019 [7] | White (fluorescent) | 32 | TMPyP | 0.5 | MS2 phage | ssRNA | 2 × 10−1 | 1 × 10−2 |
| This study | Blue (LED, 405 nm) | 60 | TMPyP | 10 | ΦX174 phage | ssDNA | 4 × 10−3 | 7 × 10−6 |
| P22 phage | dsDNA | 4 × 10−2 | 6 × 10−5 | |||||
| fr phage | ssRNA | 5 × 10−1 | 8 × 10−4 | |||||
| PdT4 | ΦX174 phage | ssDNA | 1 × 10−2 | 2 × 10−5 | ||||
| P22 phage | dsDNA | 1 × 10−1 | 2 × 10−4 | |||||
| C14 | ΦX174 phage | ssDNA ssDNA | 8 × 10−3 | 1 × 10−5 | ||||
| PdC14 | 1 × 10−2 | 2 × 10−5 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heffron, J.; Bork, M.; Mayer, B.K.; Skwor, T. A Comparison of Porphyrin Photosensitizers in Photodynamic Inactivation of RNA and DNA Bacteriophages. Viruses 2021, 13, 530. https://0-doi-org.brum.beds.ac.uk/10.3390/v13030530
Heffron J, Bork M, Mayer BK, Skwor T. A Comparison of Porphyrin Photosensitizers in Photodynamic Inactivation of RNA and DNA Bacteriophages. Viruses. 2021; 13(3):530. https://0-doi-org.brum.beds.ac.uk/10.3390/v13030530
Chicago/Turabian StyleHeffron, Joe, Matthew Bork, Brooke K. Mayer, and Troy Skwor. 2021. "A Comparison of Porphyrin Photosensitizers in Photodynamic Inactivation of RNA and DNA Bacteriophages" Viruses 13, no. 3: 530. https://0-doi-org.brum.beds.ac.uk/10.3390/v13030530