Cross-Reactivity of Human, Wild Boar, and Farm Animal Sera from Pre- and Post-Pandemic Periods with Alpha- and Βeta-Coronaviruses (CoV), including SARS-CoV-2
Abstract
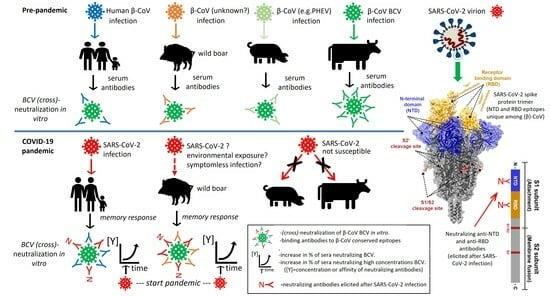
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Pre-Treatment of Sera
2.3. Immune Staining of Infected Cells
2.4. Neutralization Assays
2.4.1. M96 SARS-CoV-2 VNT
2.4.2. BCV, SARS-CoV-2 and TGEV M24 Neutralization Assays
2.4.3. Determination of the Neutralization Index (NI)
2.4.4. Pseudovirus Neutralization Assay
2.5. SARS-CoV-2 NP and S ELISA
2.6. Farm Animal and Human Sera Used for Immune Staining, Neutralization Tests and ELISAs
2.7. Statistical Analysis
3. Results
3.1. Rabbit Antibodies Raised against S2 Peptide React with Heterologous α- and β-CoVs In Vitro but Are Non-Neutralizing
3.2. Rabbit and Human Sera Stain Cells Infected with Heterologous β-CoV, including SARS-CoV-2
3.3. Human Pre-Pandemic and SARS-CoV-2-Convalescent Sera Neutralize the Heterologous β-CoV BCV In Vitro but Not the α-CoV TGEV
3.4. Testing of Panels of Pre- and Post-Pandemic Human and Animal Sera for Cross-Reactivity with CoV
3.5. Reactivity of Sera in SARS-CoV-2 NP and S ELISAs
3.6. BCV-Neutralizing Animal Sera Do Not Neutralize SARS-CoV-2 in a Standard M96 SARS-CoV-2 VNT Test
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Tomris, I.; Unione, L.; Nguyen, L.; Zaree, P.; Bouwman, K.M.; Liu, L.; Li, Z.; Fok, J.A.; Carrasco, M.R.; van der Woude, R.; et al. SARS-CoV-2 Spike N-Terminal Domain Engages 9-O-Acetylated α2–8-Linked Sialic Acids. ACS Chem. Biol. 2023, 18, 1180–1191. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- Winger, A.; Caspari, T. The Spike of Concern—The Novel Variants of SARS-CoV-2. Viruses 2021, 13, 1002. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Lee, S.-S. A Detailed Overview of Immune Escape, Antibody Escape, Partial Vaccine Escape of SARS-CoV-2 and Their Emerging Variants with Escape Mutations. Front. Immunol. 2022, 13, 801522. [Google Scholar] [CrossRef]
- Chatterjee, S.; Bhattacharya, M.; Nag, S.; Dhama, K.; Chakraborty, C. A Detailed Overview of SARS-CoV-2 Omicron: Its Sub-Variants, Mutations and Pathophysiology, Clinical Characteristics, Immunological Landscape, Immune Escape, and Therapies. Viruses 2023, 15, 167. [Google Scholar] [CrossRef]
- Shrwani, K.; Sharma, R.; Krishnan, M.; Jones, T.; Mayora-Neto, M.; Cantoni, D.; Temperton, N.J.; Dobson, S.L.; Subramaniam, K.; McNamara, P.S.; et al. Detection of Serum Cross-Reactive Antibodies and Memory Response to SARS-CoV-2 in Prepandemic and Post–COVID-19 Convalescent Samples. J. Infect. Dis. 2021, 224, 1305–1315. [Google Scholar] [CrossRef]
- Woudenberg, T.; Pelleau, S.; Anna, F.; Attia, M.; Donnadieu, F.; Gravet, A.; Lohmann, C.; Seraphin, H.; Guiheneuf, R.; Delamare, C.; et al. Humoral immunity to SARS-CoV-2 and seasonal coronaviruses in children and adults in north-eastern France. EBioMedicine 2021, 70, 103495. [Google Scholar] [CrossRef]
- Ansari, A.; Arya, R.; Sachan, S.; Jha, S.N.; Kalia, A.; Lall, A.; Sette, A.; Grifoni, A.; Weiskopf, D.; Coshic, P.; et al. Immune Memory in Mild COVID-19 Patients and Unexposed Donors Reveals Persistent T Cell Responses After SARS-CoV-2 Infection. Front. Immunol. 2021, 12, 636768. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef]
- Bianchini, F.; Crivelli, V.; Abernathy, M.E.; Guerra, C.; Palus, M.; Muri, J.; Marcotte, H.; Piralla, A.; Pedotti, M.; De Gasparo, R.; et al. Human neutralizing antibodies to cold linear epitopes and subdomain 1 of the SARS-CoV-2 spike glycoprotein. Sci. Immunol. 2023, 8, eade0958. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020, 25, 2001005. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2021, 602, 481–486. [Google Scholar] [CrossRef]
- Hossain, M.G.; Javed, A.; Akter, S.; Saha, S. SARS-CoV-2 host diversity: An update of natural infections and experimental evidence. J. Microbiol. Immunol. Infect. 2020, 54, 175–181. [Google Scholar] [CrossRef]
- Yen, H.-L.; Sit, T.H.C.; Brackman, C.J.; Chuk, S.S.Y.; Gu, H.; Tam, K.W.S.; Law, P.Y.T.; Leung, G.M.; Peiris, M.; Poon, L.L.M.; et al. Transmission of SARS-CoV-2 delta variant (AY.127) from pet hamsters to humans, leading to onward human-to-human transmission: A case study. Lancet 2022, 399, 1070–1078. [Google Scholar] [CrossRef]
- Sila, T.; Sunghan, J.; Laochareonsuk, W.; Surasombatpattana, S.; Kongkamol, C.; Ingviya, T.; Siripaitoon, P.; Kositpantawong, N.; Kanchanasuwan, S.; Hortiwakul, T.; et al. Suspected Cat-to-Human Transmission of SARS-CoV-2, Thailand, July–September 2021. Emerg. Infect. Dis. 2022, 28, 1485–1488. [Google Scholar] [CrossRef]
- EFSA Panel on Animal Health and Welfare (AHAW); Nielsen, S.S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Rojas, J.L.G.; Gortázar, C.; et al. SARS-CoV-2 in animals: Susceptibility of animal species, risk for animal and public health, monitoring, prevention and control. EFSA J. 2023, 21, e07822. [Google Scholar] [CrossRef]
- Mastutik, G.; Rohman, A.; I’Tishom, R.; Ruiz-Arrondo, I.; de Blas, I. Experimental and natural infections of severe acute respiratory syndrome-related coronavirus 2 in pets and wild and farm animals. Vet. World 2022, 15, 565–589. [Google Scholar] [CrossRef]
- Sikkema, R.S.; Tobias, T.; Oreshkova, N.; de Bruin, E.; Okba, N.; Chandler, F.; Hulst, M.M.; Rodon, J.; Houben, M.; van Maanen, K.; et al. Experimental and field investigations of exposure, replication and transmission of SARS-CoV-2 in pigs in the Netherlands. Emerg. Microbes Infect. 2021, 11, 91–94. [Google Scholar] [CrossRef]
- Ellis, J.; Sniatynski, M.; Rapin, N.; Lacoste, S.; Erickson, N.; Haines, D. SARS coronavirus 2-reactive antibodies in bovine colostrum. Can. Vet. J. 2023, 64, 337–343. [Google Scholar]
- Turlewicz-Podbielska, H.; Pomorska-Mól, M. Porcine Coronaviruses: Overview of the State of the Art. Virol. Sin. 2021, 36, 833–851. [Google Scholar] [CrossRef]
- Vijgen, L.; Keyaerts, E.; Lemey, P.; Maes, P.; Van Reeth, K.; Nauwynck, H.; Pensaert, M.; Van Ranst, M. Evolutionary History of the Closely Related Group 2 Coronaviruses: Porcine Hemagglutinating Encephalomyelitis Virus, Bovine Coronavirus, and Human Coronavirus OC43. J. Virol. 2006, 80, 7270–7274. [Google Scholar] [CrossRef]
- Schwegmann-Wessels, C.; Herrler, G. Transmissible gastroenteritis virus infection: A vanishing specter. DTW. Dtsch. Tierarztl. Wochenschr. 2006, 113, 157–159. [Google Scholar]
- Kasza, L.; Shadduck, J.A.; Christofinis, G.J. Establishment, Viral Susceptibility and Biological Characteristics of a Swine Kidney Cell Line SK-6. Res. Vet. Sci. 1972, 13, 46–53. [Google Scholar] [CrossRef]
- Liu, L.; Hägglund, S.; Hakhverdyan, M.; Alenius, S.; Larsen, L.E.; Belák, S. Molecular Epidemiology of Bovine Coronavirus on the Basis of Comparative Analyses of the S Gene. J. Clin. Microbiol. 2006, 44, 957–960. [Google Scholar] [CrossRef]
- Gerhards, N.M.; Cornelissen, J.B.W.J.; van Keulen, L.J.M.; Harders-Westerveen, J.; Vloet, R.; Smid, B.; Vastenhouw, S.; van Oort, S.; der Honing, R.W.H.-V.; Gonzales, J.L.; et al. Predictive Value of Precision-Cut Lung Slices for the Susceptibility of Three Animal Species for SARS-CoV-2 and Validation in a Refined Hamster Model. Pathogens 2021, 10, 824. [Google Scholar] [CrossRef]
- Kleine-Weber, H.; Elzayat, M.T.; Wang, L.; Graham, B.S.; Müller, M.A.; Drosten, C.; Pöhlmann, S.; Hoffmann, M. Mutations in the Spike Protein of Middle East Respiratory Syndrome Coronavirus Transmitted in Korea Increase Resistance to Antibody-Mediated Neutralization. J. Virol. 2019, 93, e01381-18. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- de Cock, M.P.; de Vries, A.; Fonville, M.; Esser, H.J.; Mehl, C.; Ulrich, R.G.; Joeres, M.; Hoffmann, D.; Eisenberg, T.; Schmidt, K.; et al. Increased rat-borne zoonotic disease hazard in greener urban areas. Sci. Total Environ. 2023, 896, 165069. [Google Scholar] [CrossRef]
- Van Zaane, D.; Hulst, M.M. Monoclonal antibodies against porcine immunoglobulin isotypes. Vet. Immunol. Immunopathol. 1987, 16, 23–36. [Google Scholar] [CrossRef]
- Hartman, H.; Wang, Y.; Schroeder, H.W., Jr.; Cui, X. Absorbance summation: A novel approach for analyzing high-throughput ELISA data in the absence of a standard. PLoS ONE 2018, 13, e0198528. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2019, 25, 2000045. [Google Scholar] [CrossRef]
- Chmielewska, A.M.; Czarnota, A.; Bieńkowska-Szewczyk, K.; Grzyb, K. Immune response against SARS-CoV-2 variants: The role of neutralization assays. NPJ Vaccines 2021, 6, 142. [Google Scholar] [CrossRef]
- Kumar, S.; Karuppanan, K.; Subramaniam, G. Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment. J. Med. Virol. 2022, 94, 4780–4791. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef]
- Ladner, J.T.; Henson, S.N.; Boyle, A.S.; Engelbrektson, A.L.; Fink, Z.W.; Rahee, F.; D’ambrozio, J.; Schaecher, K.E.; Stone, M.; Dong, W.; et al. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep. Med. 2021, 2, 100189. [Google Scholar] [CrossRef]
- Ausserwöger, H.; Schneider, M.M.; Herling, T.W.; Arosio, P.; Invernizzi, G.; Knowles, T.P.J.; Lorenzen, N. Non-specificity as the sticky problem in therapeutic antibody development. Nat. Rev. Chem. 2022, 6, 844–861. [Google Scholar] [CrossRef]
- Vogt, R.V.; Phillips, D.L.; Henderson, L.O.; Whitfield, W.; Spierto, F.W. Quantitative differences among various proteins as blocking agents for ELISA microtiter plates. J. Immunol. Methods 1987, 101, 43–50. [Google Scholar] [CrossRef]
- Nakayama, E.E.; Shioda, T. SARS-CoV-2 Related Antibody-Dependent Enhancement Phenomena In Vitro and In Vivo. Microorganisms 2023, 11, 1015. [Google Scholar] [CrossRef]
- Prévost, J.; Gasser, R.; Beaudoin-Bussières, G.; Richard, J.; Duerr, R.; Laumaea, A.; Anand, S.P.; Goyette, G.; Benlarbi, M.; Ding, S.; et al. Cross-Sectional Evaluation of Humoral Responses against SARS-CoV-2 Spike. Cell Rep. Med. 2020, 1, 100126. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e1415. [Google Scholar] [CrossRef] [PubMed]
- Lavell, A.H.A.; Sikkens, J.J.; Edridge, A.W.D.; van der Straten, K.; Sechan, F.; Oomen, M.; Buis, D.T.; Schinkel, M.; Burger, J.A.; Poniman, M.; et al. Recent infection with HCoV-OC43 may be associated with protection against SARS-CoV-2 infection. iScience 2022, 25, 105105. [Google Scholar] [CrossRef] [PubMed]
- Laidlaw, B.J.; Ellebedy, A.H. The germinal centre B cell response to SARS-CoV-2. Nat. Rev. Immunol. 2021, 22, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Lan, J.; Huang, B.; Ruhan, A.; Lu, M.; Wang, W.; Wang, W.; Li, W.; Deng, Y.; Wong, G.; et al. Lack of antibody-mediated cross-protection between SARS-CoV-2 and SARS-CoV infections. EBioMedicine 2020, 58, 102890. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Tortorici, M.A.; Frenz, B.; Snijder, J.; Li, W.; Rey, F.A.; DiMaio, F.; Bosch, B.-J.; Veesler, D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016, 23, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Lorusso, A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet. Microbiol. 2020, 244, 108693. [Google Scholar] [CrossRef]
- Lima, L.B.; Mesquita, F.P.; de Oliveira, L.L.B.; Oliveira, F.A.d.S.; de Moraes, M.E.A.; Souza, P.F.N.; Montenegro, R.C. True or false: What are the factors that influence COVID-19 diagnosis by RT-qPCR? Expert Rev. Mol. Diagn. 2022, 22, 157–167. [Google Scholar] [CrossRef]
- Jemeršić, L.; Lojkić, I.; Krešić, N.; Keros, T.; Zelenika, T.A.; Jurinović, L.; Skok, D.; Bata, I.; Boras, J.; Habrun, B.; et al. Investigating the Presence of SARS CoV-2 in Free-Living and Captive Animals. Pathogens 2021, 10, 635. [Google Scholar] [CrossRef]
- Pekar, J.E.; Magee, A.; Parker, E.; Moshiri, N.; Izhikevich, K.; Havens, J.L.; Gangavarapu, K.; Serrano, L.M.M.; Crits-Christoph, A.; Matteson, N.L.; et al. The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2. Science 2022, 377, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, A.S.; Ando, A.W.; Loch-Temzelides, T.; Vale, M.M.; Li, B.V.; Li, H.; Busch, J.; Chapman, C.A.; Kinnaird, M.; Nowak, K.; et al. The costs and benefits of primary prevention of zoonotic pandemics. Sci. Adv. 2022, 8, eabl4183. [Google Scholar] [CrossRef] [PubMed]






| Species and Origin Field Sera & | Year Collected (Pre- or Post-Pandemic) | BCV Neutralization Index (NI) | TGEV Neutralization Index (NI) | PHEV Immune Staining | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number Tested | Number Positive (%) $ | 5000 | 500 | 50 | Number Tested | Number Positive (%) $ | 10,000 | 1000 | 100 | Number Tested (BCV+) | Number Positive (%) $ | 1:750 ** [++] | 1:250 ** [+] | ||
| Goat | |||||||||||||||
| G (2 farms NL *) | 2012 (pre) | 20 | 0 | 0 | 0 | 0 | NT | − | − | − | − | NT | − | − | − |
| Rabbit | |||||||||||||||
| R-R (regular husbandry 5–7 wks) | 2019-21 (pre-post) | 20 | 12 (60%) | 4 | 1 | 7 | 5 (5) | 0 | 0 | 0 | 0 | 4 (4) | 3 | 2 | 1 |
| Cow | |||||||||||||||
| C-M (dairy cows 3 farms NL) | 2010 (pre) | 16 | 16 (100%) | 6 | 5 | 5 | 9 (9) | 0 | 0 | 0 | 0 | 5 (4) | 2 | 1 | 1 |
| Sheep | |||||||||||||||
| S-H herd 1 (NL) | 2012 (pre) | 4 | 4 | 1 | 2 | 1 | 4 (4) | 0 | 0 | 0 | 0 | 4 (4) | 0 | 0 | 0 |
| S-H herd 2 (NL) | 2012 (pre) | 2 | 0 | 0 | 0 | 0 | NT | − | − | − | − | 2 (0) | 0 | 0 | 0 |
| S-H herd 3 (NL) | 2012 (pre) | 5 | 0 | 0 | 0 | 0 | NT | − | − | − | − | NT | − | − | − |
| S-SPF (SPF herd France) | 2012 (pre) | 9 | 0 | 0 | 0 | 0 | NT | − | − | − | − | NT | − | − | − |
| Pig | |||||||||||||||
| P-F (finishing pigs NL) | 2007 (pre) | 42 | 19 (45%) | 2 | 12 | 5 | 37 (18) | 21 (57%) | 7 | 9 | 5 | 40 (17) | 11 (28%) | 2 | 9 |
| P-Exp (10–12 wks old piglets NL) | 2006 (pre) | 9 | 2 | 0 | 1 | 1 | 9 (2 ) | 9 (100%) | 1 | 5 | 3 | 9 (2) | 4 | 1 | 3 |
| P-S ([pregnant-]sows farm NL) | 2021 (post) | 6 | 1 | 0 | 0 | 1 | 6 (1) | 3 | 2 | 1 | 0 | 2 (1) | 0 | 0 | 0 |
| P-P (weaned piglets farm NL) | 2021 (post) | 21 | 0 (0%) | 0 | 0 | 0 | 21 (0) | 10 (48%) | 1 | 2 | 7 | 3 (0) | 0 | 0 | 0 |
| P-FA (fattening pigs 2 farms NL) | 2021 (post) | 56 | 18 (32%) | 7 | 6 | 5 | 28 (18) | 19 (68%) | 2 | 11 | 6 | NT | − | − | − |
| Wild Boar (all ages) | |||||||||||||||
| WB-V and WB-P (Veluwe and Peel area NL) | 2018 (pre) | 91 | 22 (24%) | 6 | 5 | 11 | 53 (16) | 2 (3.8%) | 0 | 2 | 0 | 19 (6) | 2 (11%) | 1 | 1 |
| WB-V and WB-P (Veluwe and Peel area NL) | 2021-22 (post) | 185 | 63 (34%) | 31 | 20 | 12 | 75 (59) | 4 (5.3%) | 1 | 2 | 1 | 16 (16) | 9 (56%) | 2 | 7 |
| Human | |||||||||||||||
| H-ETZ-F (pig farmers) | 2018 (pre) | 22 | 15 (68%) | 1 | 4 | 10 | 12 (8) | 0 (0%) | 0 | 0 | 0 | 15 (15) | 8 (53%) | 0 | 8 |
| H-ETZ-P (random patients) | 2018 (pre) | 30 | 11 (37%) | 4 | 2 | 5 | 12 (6) | 0 (0%) | 0 | 0 | 0 | 11 (11) | 4 (36%) | 0 | 4 |
| H-ETZ-IC, H-ETZ-W and H-WBVR (SARS-CoV-2 PCR+) | 2020 (post) | 78 | 61 (78%) | 36 | 14 | 11 | 14 (9 ) | 0 (0%) | 0 | 0 | 0 | 78 (61) | 60 (77%) | 38 | 22 |
| Species and Origin Field Sera & | Pre- or Post-Pandemic | Number (n) of Sera | BCV NI * (1:50 Dilution) | SARS-CoV-2 VNT Titer **$ (M96-Test) |
|---|---|---|---|---|
| Hamster | ||||
| HAM-d0 (pre-serum before infection) | (−) control | 1 | 0 | <10 |
| HAM-d21 (SARS-CoV-2 infected) | (+) control | 1 | 0 | 250 |
| Rabbit | ||||
| R-R (regular husbandry 5–7 wks) | pre- | 2 | 50|5000 | <10 (n = 2) |
| Cow | ||||
| C-M (dairy cows NL) | pre- | 11 | 5000 (n = 11) | <10 (n = 11) |
| Pig | ||||
| P-F (finishing pigs NL) | pre- | 2 | 500|5000 | <10 (n = 2) |
| P-Exp (10–12 wks old piglets NL) | pre- | 1 | 0 | <10 (n = 1) |
| P-S ([pregnant-]sows farm NL) | post | 1 | 50 | <10 (n = 1) |
| Wild Boar | ||||
| WB-V (Veluwe region NL) | post | 6 | 5000 (n = 6) | <10 (n = 3)|10, 30, 50 (n = 3) |
| WB-P (Peel region NL) | post | 8 | 500 (n = 1)|5000 (n = 7) | <10 (n = 6)|10, 15 (n = 2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hulst, M.; Kant, A.; Harders-Westerveen, J.; Hoffmann, M.; Xie, Y.; Laheij, C.; Murk, J.-L.; Van der Poel, W.H.M. Cross-Reactivity of Human, Wild Boar, and Farm Animal Sera from Pre- and Post-Pandemic Periods with Alpha- and Βeta-Coronaviruses (CoV), including SARS-CoV-2. Viruses 2024, 16, 34. https://0-doi-org.brum.beds.ac.uk/10.3390/v16010034
Hulst M, Kant A, Harders-Westerveen J, Hoffmann M, Xie Y, Laheij C, Murk J-L, Van der Poel WHM. Cross-Reactivity of Human, Wild Boar, and Farm Animal Sera from Pre- and Post-Pandemic Periods with Alpha- and Βeta-Coronaviruses (CoV), including SARS-CoV-2. Viruses. 2024; 16(1):34. https://0-doi-org.brum.beds.ac.uk/10.3390/v16010034
Chicago/Turabian StyleHulst, Marcel, Arie Kant, José Harders-Westerveen, Markus Hoffmann, Yajing Xie, Charlotte Laheij, Jean-Luc Murk, and Wim H. M. Van der Poel. 2024. "Cross-Reactivity of Human, Wild Boar, and Farm Animal Sera from Pre- and Post-Pandemic Periods with Alpha- and Βeta-Coronaviruses (CoV), including SARS-CoV-2" Viruses 16, no. 1: 34. https://0-doi-org.brum.beds.ac.uk/10.3390/v16010034