In Silico Structural Evaluation of Short Cationic Antimicrobial Peptides
Abstract
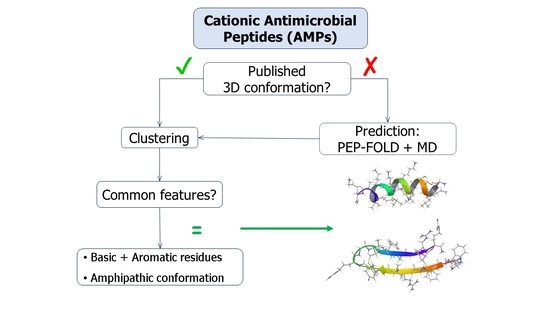
:1. Introduction
2. Materials and Methods
2.1. Data Collation
2.2. Structure Prediction and Molecular Dynamics Simulation
2.3. Hierarchical Clustering
2.4. Molecular Dynamics Simulation of Peptides in the Presence of Micelles
3. Results
3.1. Investigation of a Set of Antimicrobial Peptides with Known Confomation
3.2. Investigation of a Set of Antimicrobial Peptides with No Known Conformation
3.3. A System to Study the Interaction of Short Antimicrobial Peptides with a Membrane-Like Structure
4. Discussion
4.1. Investigation of a Set of Antimicrobial Peptides with Known Confomations
4.2. Evaluation of In Silico Appraoches to Predict Conformation of Short Cationic Peptides
4.3. Common Structural Features of Antimicrobial Peptides with Activity against Gram Negative Bacteria
4.4. MD Simulation to Evaluate Interaction of Short Antimicrobial Peptides with a Membrane-Like Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, R.; Liu, Y.; Zhang, Q.; Jin, L.; Wang, Q.; Zhang, Y.; Wang, X.; Hu, M.; Li, L.; Qi, J.; et al. The prevalence of colistin resistance in escherichia coli and klebsiella pneumoniae isolated from food animals in china: Coexistence of mcr-1 and blandm with low fitness cost. Int. J. Antimicrob. Agents 2018, 51, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Bechinger, B.; Gorr, S.U. Antimicrobial peptides: Mechanisms of action and resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Thevenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tuffery, P. Pep-fold: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef] [PubMed]
- Mikut, R.; Ruden, S.; Reischl, M.; Breitling, F.; Volkmer, R.; Hilpert, K. Improving short antimicrobial peptides despite elusive rules for activity. Biochim. Biophys. Acta 2016, 1858, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Marquette, A.; Bechinger, B. Biophysical investigations elucidating the mechanisms of action of antimicrobial peptides and their synergism. Biomolecules 2018, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Rangarajan, N.; Weisshaar, J.C. Lights, camera, action! Antimicrobial peptide mechanisms imaged in space and time. Trends Microbiol. 2016, 24, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, T.; Wei, D.; Strandberg, E.; Ulrich, A.S.; Ulmschneider, J.P. How reliable are molecular dynamics simulations of membrane active antimicrobial peptides? Biochim. Biophys. Acta-Biomembr. 2014, 1838, 2280–2288. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank nucleic acids research. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Uggerhoj, L.E.; Poulsen, T.J.; Munk, J.K.; Fredborg, M.; Sondergaard, T.E.; Frimodt-Moller, N.; Hansen, P.R.; Wimmer, R. Rational design of alpha-helical antimicrobial peptides: Do’s and don’ts. Chembiochem 2015, 16, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Rozek, A.; Friedrich, C.L.; Hancock, R.E. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry 2000, 39, 15765–15774. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Peterkofsky, A.; Wang, G. Nmr studies of aurein 1.2 analogs. Biochim. Biophys. Acta 2006, 1758, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Hori, Y.; Demura, M.; Iwadate, M.; Ulrich, A.S.; Niidome, T.; Aoyagi, H.; Asakura, T. Interaction of mastoparan with membranes studied by 1h-nmr spectroscopy in detergent micelles and by solid-state 2h-nmr and 15n-nmr spectroscopy in oriented lipid bilayers. Eur. J. Biochem. 2001, 268, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Tinoco, L.W.; Gomes-Neto, F.; Valente, A.P.; Almeida, F.C. Effect of micelle interface on the binding of anticoccidial pw2 peptide. J. Biomol. NMR 2007, 39, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Respondek, M.; Madl, T.; Gobl, C.; Golser, R.; Zangger, K. Mapping the orientation of helices in micelle-bound peptides by paramagnetic relaxation waves. J. Am. Chem. Soc. 2007, 129, 5228–5234. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, R.; Joshi, M.; Mohanram, H.; Bhunia, A.; Mangoni, M.L.; Bhattacharjya, S. NMR structure of temporin-1 Ta in lipopolysaccharide micelles: Mechanistic insight into inactivation by outer membrane. PLoS ONE 2013, 8, e72718. [Google Scholar] [CrossRef] [PubMed]
- Schibli, D.J.; Hwang, P.M.; Vogel, H.J. Structure of the antimicrobial peptide tritrpticin bound to micelles: A distinct membrane-bound peptide fold. Biochemistry 1999, 38, 16749–16755. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Shin, S.Y.; Kim, K.; Lim, S.S.; Hahm, K.S.; Kim, Y. Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide isct and its analogs. Biochem. Biophys. Res. Commun. 2004, 323, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Aumelas, A.; Gao, B. Convergent evolution-guided design of antimicrobial peptides derived from influenza a virus hemagglutinin. J. Med. Chem. 2011, 54, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.E.R.; Fadel, V.; Maltarollo, V.G.; Baldissera, G.; Honorio, K.M.; Ruggiero, J.R.; Dos Santos Cabrera, M.P. Md simulations and multivariate studies for modeling the antileishmanial activity of peptides. Chem. Biol. Drug Dis. 2017, 90, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Gunaskekera, S.; Muhammad, T.; Stromstedt, A.A.; Rosengren, K.J.; Goransson, U. Backbone-Cyclized Stable Peptide-Dimers Derived from the Human Cathelicidin ll-37 Mediate Potent Antimicrobial Activity. Available online: https://www.rcsb.org/structure/2NA3 (accessed on 19 June 2018).
- Wang, G.; Li, X.; Wang, Z. Apd3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 84. [Google Scholar]
- Kaminski, G.A.; Friesner, R.A. Evaluation and reparametrization of the opls-aa force field for proteins via comparison with accurate quantum chemical calculations of peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Herbert, A.; Sternberg, M. Maxcluster—A Tool for Protein Structure Comparison and Clustering. 2014. Available online: http://www.sbg.bio.ic.ac.uk/~maxcluster/ (accessed on 19 June 2018).
- Konno, K.; Hisada, M.; Fontana, R.; Lorenzi, C.C.B.; Naoki, H.; Itagaki, Y.; Miwa, A.; Kawai, N.; Nakata, Y.; Yashuhara, T.; et al. Anoplin, a novel antimicrobial peptide from the venom of the solitary wasp anoplius samariensis. Biochim. Biophys. Acta 2001, 1550, 70–80. [Google Scholar] [CrossRef]
- Martinez, L.; Andrade, R.; Birgin, E.G.; Martinez, J.M. Packmol: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. 1976, 32, 922–923. [Google Scholar] [CrossRef] [Green Version]
- Kabsch, W. A discussion of the solution for the best rotation to relate two sets of vectors. Acta Crystallogr. 1978, 34, 827–828. [Google Scholar] [CrossRef] [Green Version]
- Nesuta, O.; Budesinsky, M.; Hadravova, R.; Monincova, L.; Humpolickova, J.; Cerovsky, V. How proteases from enterococcus faecalis contribute to its resistance to short alpha-helical antimicrobial peptides. Pathog. Dis. 2017, 75. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W. Peptide antibiotics. Lancet 1997, 349, 418–422. [Google Scholar] [CrossRef]
- Michalek, M.; Jung, S.; Shomali, M.R.; Cauchard, S.; Sonnichsen, F.D.; Grotzinger, J. Solution structure and functional studies of the highly potent equine antimicrobial peptide defa1. Biochem. Biophys. Res. Commun. 2015, 459, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Kushibiki, T.; Kamiya, M.; Aizawa, T.; Kumaki, Y.; Kikukawa, T.; Mizuguchi, M.; Demura, M.; Kawabata, S.; Kawano, K. Interaction between tachyplesin i, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide. Biochim. Biophys. Acta 2014, 1844, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Beaufays, J.; Lins, L.; Thomas, A.; Brasseur, R. In silico predictions of 3d structures of linear and cyclic peptides with natural and non-proteinogenic residues. J. Pept. Sci. 2012, 18, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Amitay, M.; Goldstein, M. Evaluating the peptide structure prediction capabilities of a purely ab-initio method. Protein Eng. Des. Sel. 2017, 30, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Quintero-Hernández, V.; Vargas-Jaimes, L.; Batista, C.V.; Winkel, K.D.; Possani, L.D. Characterization of the venom from the australian scorpion urodacus yaschenkoi: Molecular mass analysis of components, cdna sequences and peptides with antimicrobial activity. Toxicon 2013, 63, 44–45. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Cabrera, M.P.; Arcisio-Miranda, M.; Broggio Costa, S.T.; Konno, K.; Ruggiero, J.R.; Procopio, J.; Ruggiero Neto, J. Study of the mechanism of action of anoplin, a helical antimicrobial decapeptide with ion channel-like activity, and the role of the amidated c-terminus. J. Pept. Sci. 2008, 14, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, H.; Tuknait, A.; Chaudhary, K.; Singh, B.; Kumaran, S.; Raghava, G.P.S. Pepstrmod: Structure prediction of peptides containing natural, non-natural and modified residues. Boil. Direct 2015, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Nguyen, P.H.; Nasica-Labouze, J.; Mu, Y.; Derreumaux, P. Folding atomistic proteins in explicit solvent using simulated tempering. J. Phys. Chem. B 2015, 119, 6941–6951. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.-F.; Xu, W.-F.; Yang, S.-G.; Yang, G.-F. Multiple simulated annealing-molecular dynamics (msa-md) for conformational space search of peptide and miniprotein. Sci. Rep. 2015, 5, 15568. [Google Scholar] [CrossRef] [PubMed]
- London, N.; Raveh, B.; Cohen, E.; Fathi, G.; Schueler-Furman, O. Rosetta flexpepdock web server--high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011, 39, W249–W253. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, T.J. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1999, 181, 4725–4733. [Google Scholar] [PubMed]
- Dubovskii, P.V.; Vassilevski, A.A.; Samsonova, O.V.; Egorova, N.S.; Kozlov, S.A.; Feofanov, A.V.; Arseniev, A.S.; Grishin, E.V. Novel lynx spider toxin shares common molecular architecture with defense peptides from frog skin. FEBS J. 2011, 278, 4382–4393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillong, M.; Hiss, J.A.; Schneider, P.; Lin, Y.C.; Posselt, G.; Pfeiffer, B.; Blatter, M.; Muller, A.T.; Bachler, S.; Neuhaus, C.S.; et al. Rational design of membrane-pore-forming peptides. Small 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Usachev, K.S.; Kolosova, O.A.; Klochkova, E.A.; Yulmetov, A.R.; Aganov, A.V.; Klochkov, V.V. Oligomerization of the antimicrobial peptide protegrin-5 in a membrane-mimicking environment. Structural studies by high-resolution nmr spectroscopy. Eur. Biophys. J. 2017, 46, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Pal, I.; Atreya, H.S.; Bhunia, A. Antimicrobial Peptide Andersonin-y1 (ay1). Available online: http://www.rcsb.org/structure/5YKK (accessed on 14 May 2018).
- Herrera, A.; Xiao, Y.; Soulages, J.L.; Bommineni, Y.R.; Prakash, O.; Zhang, G. The Central Kink Region of Alpha-Helical Fowlicidin-2 Is Critically Involved in Antibacterial and Lipopolysaccharide-Neutralizing Activities. Available online: http://www.rcsb.org/structure/2GDL (accessed on 14 May 2018).
- Bommineni, Y.R.; Dai, H.; Gong, Y.X.; Soulages, J.L.; Fernando, S.C.; Desilva, U.; Prakash, O.; Zhang, G. Fowlicidin-3 is an alpha-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities. FEBS J. 2007, 274, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Vassilevski, A.A.; Musolyamov, A.; Nikonorova, A.; Finikina, E.; Slavokhotova, A.; Shagin, D.; Bozin, T.N.; BOcharov, E.V.; Ovchinnikova, T.; Arseniev, A.A.; et al. Common Chickweed (Stellaria Media) Antifungal Peptides with Chitin-Binding Domain Provide Unique Plant Defense Strategy. Available online: http://www.rcsb.org/structure/2KUS (accessed on 14 May 2018).
- Shenkarev, Z.O.; Panteleev, P.V.; Balandin, S.V.; Gizatullina, A.K.; Altukhov, D.A.; Finkina, E.I.; Kokryakov, V.N.; Arseniev, A.S.; Ovchinnikova, T.V. Recombinant expression and solution structure of antimicrobial peptide aurelin from jellyfish aurelia aurita. Biochem. Biophys. Res. Commun. 2012, 429, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Wommack, A.J.; Robson, S.A.; Wanniarachchi, Y.A.; Wan, A.; Turner, C.J.; Wagner, G.; Nolan, E.M. Nmr solution structure and condition-dependent oligomerization of the antimicrobial peptide human defensin 5. Biochemistry 2012, 51, 9624–9637. [Google Scholar] [CrossRef] [PubMed]
- Conibear, A.C.; Rosengren, K.J.; Daly, N.L.; Henriques, S.T.; Craik, D.J. The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity. J. Boil. Chem. 2013, 288, 10830–10840. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikov, K.V.; Kristiansen, P.E.; Uzelac, G.; Topisirovic, L.; Kojic, M.; Nissen-Meyer, J.; Nes, I.F.; Diep, D.B. Defining the structure and receptor binding domain of the leaderless bacteriocin lsbb. J. Boil. Chem. 2014, 289, 23838–23845. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Wang, G.; Tam, Y.T.; Tam, C. Membrane-active epithelial keratin 6a fragments (kamps) are unique human antimicrobial peptides with a non-alphabeta structure. Front. Microbiol. 2016, 7, 1799. [Google Scholar] [CrossRef] [PubMed]
- Mandard, N.; Bulet, P.; Caille, A.; Daffre, S.; Vovelle, F. The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider. Eur. J. Biochem. 2002, 269, 1190–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourbigot, S.; Dodd, E.; Horwood, C.; Cumby, N.; Fardy, L.; Welch, W.H.; Ramjan, Z.; Sharma, S.; Waring, A.J.; Yeaman, M.R.; et al. Antimicrobial peptide rp-1 structure and interactions with anionic versus zwitterionic micelles. Biopolymers 2009, 91, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dubovskii, P.V.; Volynsky, P.E.; Polyansky, A.A.; Karpunin, D.V.; Chupin, V.V.; Efremov, R.G.; Arseniev, A.S. Three-dimensional structure/hydrophobicity of latarcins specifies their mode of membrane activity. Biochemistry 2008, 47, 3525–3533. [Google Scholar] [CrossRef] [PubMed]
- Petit, V.W.; Rolland, J.L.; Blond, A.; Cazevieille, C.; Djediat, C.; Peduzzi, J.; Goulard, C.; Bachere, E.; Dupont, J.; Destoumieux-Garzon, D.; et al. A hemocyanin-derived antimicrobial peptide from the penaeid shrimp adopts an alpha-helical structure that specifically permeabilizes fungal membranes. Biochim. Biophys. Acta 2016, 1860, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Aboye, T.L.; Stromstedt, A.A.; Gunasekera, S.; Bruhn, J.G.; El-Seedi, H.; Rosengren, K.J.; Goransson, U. A cactus-derived toxin-like cystine knot peptide with selective antimicrobial activity. Chembiochem 2015, 16, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Berkut, A.A.; Usmanova, D.R.; Peigneur, S.; Oparin, P.B.; Mineev, K.S.; Odintsova, T.I.; Tytgat, J.; Arseniev, A.S.; Grishin, E.V.; Vassilevski, A.A. Structural similarity between defense peptide from wheat and scorpion neurotoxin permits rational functional design. J. Boil. Chem. 2014, 289, 14331–14340. [Google Scholar] [CrossRef] [PubMed]
- Subasinghage, A.P.; O’Flynn, D.; Conlon, J.M.; Hewage, C.M. Conformational and membrane interaction studies of the antimicrobial peptide alyteserin-1c and its analogue [e4k]alyteserin-1c. Biochim. Biophys. Acta 2011, 1808, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.T.; Tan, C.; Craik, D.J.; Clark, R.J. Structural and Functional Analysis of Human Liver-Expressed Antimicrobial Peptide 2. Available online: http://www.rcsb.org/structure/2L1Q (accessed on 19 June 2018).
- Silva, F.D.; Rezende, C.A.; Rossi, D.C.; Esteves, E.; Dyszy, F.H.; Schreier, S.; Gueiros-Filho, F.; Campos, C.B.; Pires, J.R.; Daffre, S. Structure and mode of action of microplusin, a copper ii-chelating antimicrobial peptide from the cattle tick rhipicephalus (boophilus) microplus. J. Boil. Chem. 2009, 284, 34735–34746. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, T.V.; Shenkarev, Z.O.; Nadezhdin, K.D.; Balandin, S.V.; Zhmak, M.N.; Kudelina, I.A.; Finkina, E.I.; Kokryakov, V.N.; Arseniev, A.S. Recombinant expression, synthesis, purification, and solution structure of arenicin. Biochem. Biophys. Res. Commun. 2007, 360, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Hilge, M.; Gloor, S.M.; Rypniewski, W.; Sauer, O.; Heightman, T.D.; Zimmermann, W.; Winterhalter, K.; Piontek, K. High-resolution native and complex structures of thermostable beta-mannanase from thermomonospora fusca—Substrate specificity in glycosyl hydrolase family 5. Structure 1998, 6, 1433–1444. [Google Scholar] [CrossRef]
- Rosenzweig, A.C.; Frederick, C.A.; Lippard, S.J.; Nordlund, P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 1993, 366, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Tack, B.F.; Sawai, M.V.; Kearney, W.R.; Robertson, A.D.; Sherman, M.A.; Wang, W.; Hong, T.; Boo, L.M.; Wu, H.; Waring, A.J.; et al. Smap-29 has two lps-binding sites and a central hinge. Eur. J. Biochem. 2002, 269, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Brunhuber, N.M.; Thoden, J.B.; Blanchard, J.S.; Vanhooke, J.L. Rhodococcus l-phenylalanine dehydrogenase: Kinetics, mechanism, and structural basis for catalytic specificity. Biochemistry 2000, 39, 9174–9187. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.V.; Myshkin, M.Y.; Shenkarev, Z.O.; Ovchinnikova, T.V. Dimerization of the antimicrobial peptide arenicin plays a key role in the cytotoxicity but not in the antibacterial activity. Biochem. Biophys. Res. Commun. 2017, 482, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Pal, I.; Atreya, H.S.; Bhunia, A. Antimicrobial Peptide ay1c Designed from the Skin Secretion of Chinese Odorous Frogs. Available online: http://www.rcsb.org/structure/5YKL (accessed on 14 May 2018).
- Wang, G.; Elliott, M.; Cogen, A.L.; Ezell, E.L.; Gallo, R.L.; Hancock, R.E. Structure, dynamics, and antimicrobial and immune modulatory activities of human ll-23 and its single-residue variants mutated on the basis of homologous primate cathelicidins. Biochemistry 2012, 51, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Uteng, M.; Hauge, H.H.; Markwick, P.R.; Fimland, G.; Mantzilas, D.; Nissen-Meyer, J.; Muhle-Goll, C. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin p and a sakacin p variant that is structurally stabilized by an inserted c-terminal disulfide bridge. Biochemistry 2003, 42, 11417–11426. [Google Scholar] [CrossRef] [PubMed]
- Sprules, T.; Kawulka, K.E.; Gibbs, A.C.; Wishart, D.S.; Vederas, J.C. Nmr solution structure of the precursor for carnobacteriocin b2, an antimicrobial peptide from carnobacterium piscicola. Eur. J. Biochem. 2004, 271, 1748–1756. [Google Scholar] [CrossRef] [PubMed]
- Andra, J.; Jakovkin, I.; Grotzinger, J.; Hecht, O.; Krasnosdembskaya, A.D.; Goldmann, T.; Gutsmann, T.; Leippe, M. Structure and mode of action of the antimicrobial peptide arenicin. Biochem. J. 2008, 410, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Campagna, S.; Saint, N.; Molle, G.; Aumelas, A. Structure and mechanism of action of the antimicrobial peptide piscidin. Biochemistry 2007, 46, 1771–1778. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.S.; Chen, C. Solution Structure of a Novel Antimicrobial Peptide, p1, from Jumper Ant Myrmecia Pilosula. Available online: http://www.rcsb.org/structure/5X3L (accessed on 19 June 2018).
- Nolde, S.B.; Vassilevski, A.A.; Rogozhin, E.A.; Barinov, N.A.; Balashova, T.A.; Samsonova, O.V.; Baranov, Y.V.; Feofanov, A.V.; Egorov, T.A.; Arseniev, A.S.; et al. Disulfide-stabilized helical hairpin structure and activity of a novel antifungal peptide ecamp1 from seeds of barnyard grass (echinochloa crus-galli). J. Boil. Chem. 2011, 286, 25145–25153. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, A.; Ramamoorthy, A.; Bhattacharjya, S. Helical hairpin structure of a potent antimicrobial peptide msi-594 in lipopolysaccharide micelles by nmr spectroscopy. Chemistry 2009, 15, 2036–2040. [Google Scholar] [CrossRef] [PubMed]
- Dubovskii, P.V.; Volynsky, P.E.; Polyansky, A.A.; Chupin, V.V.; Efremov, R.G.; Arseniev, A.S. Spatial structure and activity mechanism of a novel spider antimicrobial peptide. Biochemistry 2006, 45, 10759–10767. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.W.; Kim, J.S.; Kim, D.H.; Lee, S.H.; Park, Y.H.; Han, K.H. Solution structure and membrane interaction mode of an antimicrobial peptide gaegurin 4. Biochem. Biophys. Res. Commun. 2007, 352, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Shalev, D.E.; Rotem, S.; Fish, A.; Mor, A. Consequences of n-acylation on structure and membrane binding properties of dermaseptin derivative k4-s4-(1-13). J. Boil. Chem. 2006, 281, 9432–9438. [Google Scholar] [CrossRef] [PubMed]
- Fujitani, N.; Kouno, T.; Nakahara, T.; Takaya, K.; Osaki, T.; Kawabata, S.; Mizuguchi, M.; Aizawa, T.; Demura, M.; Nishimura, S.; et al. The solution structure of horseshoe crab antimicrobial peptide tachystatin b with an inhibitory cystine-knot motif. J. Pept. Sci. 2007, 13, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Agadi, N.; Kumar, A. Structure, Function and Membrane Interaction Studies of Two Synthetic Antimicrobial Peptides Using Solution and Solid State nmr. (2ndc). Available online: https://www.rcsb.org/structure/2ndc (accessed on 14 May 2018).
- Li, X.; Li, Y.; Han, H.; Miller, D.W.; Wang, G. Solution structures of human ll-37 fragments and nmr-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J. Am. Chem. Soc. 2006, 128, 5776–5785. [Google Scholar] [CrossRef] [PubMed]
- Godreuil, S.; Leban, N.; Padilla, A.; Hamel, R.; Luplertlop, N.; Chauffour, A.; Vittecoq, M.; Hoh, F.; Thomas, F.; Sougakoff, W.; et al. Aedesin: Structure and antimicrobial activity against multidrug resistant bacterial strains. PLoS ONE 2014, 9, e105441. [Google Scholar] [CrossRef] [PubMed]
- Sawai, M.V.; Waring, A.J.; Kearney, W.R.; McCray, P.B., Jr.; Forsyth, W.R.; Lehrer, R.I.; Tack, B.F. Impact of single-residue mutations on the structure and function of ovispirin/novispirin antimicrobial peptides. Protein Eng. 2002, 15, 225–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, J.P.; Rozek, A.; Hancock, R.E. Structure-activity relationships for the beta-hairpin cationic antimicrobial peptide polyphemusin i. Biochim. Biophys. Acta 2004, 1698, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Poncet, J.; Garnier, J.; Zatylny, C.; Bachere, E.; Aumelas, A. Solution structure of the recombinant penaeidin-3, a shrimp antimicrobial peptide. J. Boil. Chem. 2003, 278, 36859–36867. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, P.E.; Fimland, G.; Mantzilas, D.; Nissen-Meyer, J. Structure and mode of action of the membrane-permeabilizing antimicrobial peptide pheromone plantaricin a. J. Boil. Chem. 2005, 280, 22945–22950. [Google Scholar] [CrossRef] [PubMed]
- Syvitski, R.T.; Burton, I.; Mattatall, N.R.; Douglas, S.E.; Jakeman, D.L. Structural characterization of the antimicrobial peptide pleurocidin from winter flounder. Biochemistry 2005, 44, 7282–7293. [Google Scholar] [CrossRef] [PubMed]
- Haugen, H.S.; Fimland, G.; Nissen-Meyer, J.; Kristiansen, P.E. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide curvacin a. Biochemistry 2005, 44, 16149–16157. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; Kamiya, M.; Kushibiki, T.; Nakazumi, T.; Tomisawa, S.; Abe, C.; Kumaki, Y.; Kikukawa, T.; Demura, M.; Kawano, K.; et al. Lipopolysaccharide Bound Structure of Antimicrobial Peptide Cecropin p1 by Nmr Spectroscopy. Available online: http://www.rcsb.org/structure/2N92 (accessed on 14 May 2018).
- Hunter, H.N.; Fulton, D.B.; Ganz, T.; Vogel, H.J. The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J. Boil. Chem. 2002, 277, 37597–37603. [Google Scholar] [CrossRef] [PubMed]
- Logashina, Y.A.; Solstad, R.G.; Mineev, K.S.; Korolkova, Y.V.; Mosharova, I.V.; Dyachenko, I.A.; Palikov, V.A.; Palikova, Y.A.; Murashev, A.N.; Arseniev, A.S.; et al. New disulfide-stabilized fold provides sea anemone peptide to exhibit both antimicrobial and trpa1 potentiating properties. Toxins 2017, 9, 154. [Google Scholar] [CrossRef] [PubMed]
- Maksimov, M.O.; Pelczer, I.; Link, A.J. Precursor-centric genome-mining approach for lasso peptide discovery. Proc. Natl. Acad. Sci. USA 2012, 109, 15223–15228. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Kundu, P.; Bhunia, A. Designing potent antimicrobial peptides by disulphide linked dimerization and n-terminal lipidation to increase antimicrobial activity and membrane perturbation: Structural insights into lipopolysaccharide binding. J. Colloid Interface Sci. 2016, 461, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Suetake, T.; Tsuda, S.; Kawabata, S.; Miura, K.; Iwanaga, S.; Hikichi, K.; Nitta, K.; Kawano, K. Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif. J. Boil. Chem. 2000, 275, 17929–17932. [Google Scholar] [CrossRef] [PubMed]
- Monincova, L.; Budesinsky, M.; Cujova, S.; Cerovsky, V.; Veverka, V. Structural basis for antimicrobial activity of lasiocepsin. Chembiochem 2014, 15, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Jordan, J.B.; Poppe, L.; Haniu, M.; Arvedson, T.; Syed, R.; Li, V.; Kohno, H.; Kim, H.; Schnier, P.D.; Harvey, T.S.; et al. Hepcidin revisited, disulfide connectivity, dynamics, and structure. J. Boil. Chem. 2009, 284, 24155–24167. [Google Scholar] [CrossRef] [PubMed]
- Mohanram, H. NMR structure of temporin-1 Ta in lipopolysaccharide micelles: Mechanistic insight into inactivation by outer membrane. Available online: http://www.rcsb.org/structure/2MAA (accessed on 19 June 2018).
- Yang, S.; Jung, H.; Kim, J. Solution Structure of Bmap-27. Available online: http://www.rcsb.org/structure/2KET (accessed on 14 May 2018).
- Agadi, N.; Kumar, A. Structure, Function and Membrane Interaction Studies of Two SYNTHETIC Antimicrobial Peptides Using Solution and Solid State Nmr. (2nde). Available online: http://www.rcsb.org/structure/2NDE (accessed on 14 May 2018).
- Powers, J.P.; Tan, A.; Ramamoorthy, A.; Hancock, R.E. Solution structure and interaction of the antimicrobial polyphemusins with lipid membranes. Biochemistry 2005, 44, 15504–15513. [Google Scholar] [CrossRef] [PubMed]
- Laederach, A.; Andreotti, A.H.; Fulton, D.B. Solution and micelle-bound structures of tachyplesin i and its active aromatic linear derivatives. Biochemistry 2002, 41, 12359–12368. [Google Scholar] [CrossRef] [PubMed]
- Sit, C.S.; McKay, R.T.; Hill, C.; Ross, R.P.; Vederas, J.C. The 3d structure of thuricin cd, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links. J. Am. Chem. Soc. 2011, 133, 7680–7683. [Google Scholar] [CrossRef] [PubMed]
- Mineev, K.S.; Berkut, A.A.; Novikova, R.V.; Oparin, P.B.; Grishin, E.V.; Arseniev, A.S.; Vassilevski, A.A. Nmr Spatial Structure of Tk-Hefu Peptide. Available online: http://www.rcsb.org/structure/5LM0 (accessed on 16 May 2018).
- Metelev, M.; Arseniev, A.; Bushin, L.B.; Kuznedelov, K.; Artamonova, T.O.; Kondratenko, R.; Khodorkovskii, M.; Seyedsayamdost, M.R.; Severinov, K. Acinetodin and klebsidin, rna polymerase targeting lasso peptides produced by human isolates of acinetobacter gyllenbergii and klebsiella pneumoniae. ACS Chem. Boil. 2017, 12, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ducasse, R.; Blond, A.; Zirah, S.; Goulard, C.; Lescop, E.; Guittet, E.; Pernodet, J.; Rebuffat, S. Structure and Biosynthesis of Sviceucin, an Antibacterial Type i Lasso Peptide from Streptomyces Sviceus. Available online: http://www.rcsb.org/structure/2LS1 (accessed on 16 May 2018).
- Rogne, P.; Fimland, G.; Nissen-Meyer, J.; Kristiansen, P.E. Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin g. Biochim. Biophys. Acta 2008, 1784, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Mandard, N.; Sy, D.; Maufrais, C.; Bonmatin, J.M.; Bulet, P.; Hetru, C.; Vovelle, F. Androctonin, a novel antimicrobial peptide from scorpion androctonus australis: Solution structure and molecular dynamics simulations in the presence of a lipid monolayer. J. Biomol. Struct. Dyn. 1999, 17, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Hwang, P.M.; Zhou, N.; Shan, X.; Arrowsmith, C.H.; Vogel, H.J. Three-dimensional solution structure of lactoferricin b, an antimicrobial peptide derived from bovine lactoferrin. Biochemistry 1998, 37, 4288–4298. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Bhattacharyya, D.; Singh, S.; Ghosh, A.; Schmidtchen, A.; Malmsten, M.; Bhunia, A. Role of aromatic amino acids in lipopolysaccharide and membrane interactions of antimicrobial peptides for use in plant disease control. J. Boil. Chem. 2016, 291, 13301–13317. [Google Scholar] [CrossRef] [PubMed]
- Usachev, K.S.; Efimov, S.V.; Kolosova, O.A.; Klochkova, E.A.; Aganov, A.V.; Klochkov, V.V. Antimicrobial peptide protegrin-3 adopt an antiparallel dimer in the presence of dpc micelles: A high-resolution nmr study. J. Biomol. NMR 2015, 62, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Usachev, K.S.; Efimov, S.V.; Kolosova, O.A.; Filippov, A.V.; Klochkov, V.V. High-resolution nmr structure of the antimicrobial peptide protegrin-2 in the presence of dpc micelles. J. Biomol. NMR 2015, 61, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, R.L.; Dieckmann, T.; Harwig, S.S.; Lehrer, R.I.; Eisenberg, D.; Feigon, J. Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes. Chem. Boil. 1996, 3, 543–550. [Google Scholar] [CrossRef]
- Lohans, C.T.; Towle, K.M.; Miskolzie, M.; McKay, R.T.; van Belkum, M.J.; McMullen, L.M.; Vederas, J.C. Solution structures of the linear leaderless bacteriocins enterocin 7a and 7b resemble carnocyclin a, a circular antimicrobial peptide. Biochemistry 2013, 52, 3987–3994. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, A.; Domadia, P.N.; Torres, J.; Hallock, K.J.; Ramamoorthy, A.; Bhattacharjya, S. Nmr structure of pardaxin, a pore-forming antimicrobial peptide, in lipopolysaccharide micelles: Mechanism of outer membrane permeabilization. J. Boil. Chem. 2010, 285, 3883–3895. [Google Scholar] [CrossRef] [PubMed]
- Landon, C.; Meudal, H.; Boulanger, N.; Bulet, P.; Vovelle, F. Solution structures of stomoxyn and spinigerin, two insect antimicrobial peptides with an alpha-helical conformation. Biopolymers 2006, 81, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Liao, Y.; Chen, C. Solution Structure of Tilapia Piscidin 4, an Antimicrobial Peptide Effective against Multidrug Resistant Helicobacter Pylori. Available online: http://www.rcsb.org/structure/5H2S (accessed on 16 May 2018).
- Jung, S.; Dingley, A.J.; Augustin, R.; Anton-Erxleben, F.; Stanisak, M.; Gelhaus, C.; Gutsmann, T.; Hammer, M.U.; Podschun, R.; Bonvin, A.M.; et al. Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan hydra. J. Boil. Chem. 2009, 284, 1896–1905. [Google Scholar] [CrossRef] [PubMed]
- Mygind, P.H.; Fischer, R.L.; Schnorr, K.M.; Hansen, M.T.; Sonksen, C.P.; Ludvigsen, S.; Raventos, D.; Buskov, S.; Christensen, B.; De Maria, L.; et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 2005, 437, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Rogne, P.; Haugen, C.; Fimland, G.; Nissen-Meyer, J.; Kristiansen, P.E. Three-dimensional structure of the two-peptide bacteriocin plantaricin jk. Peptides 2009, 30, 1613–1621. [Google Scholar] [CrossRef] [PubMed]
- Japelj, B.; Zorko, M.; Majerle, A.; Pristovsek, P.; Sanchez-Gomez, S.; Martinez de Tejada, G.; Moriyon, I.; Blondelle, S.E.; Brandenburg, K.; Andra, J.; et al. The acyl group as the central element of the structural organization of antimicrobial lipopeptide. J. Am. Chem. Soc. 2007, 129, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Resende, J.M.; Moraes, C.M.; Prates, M.V.; Cesar, A.; Almeida, F.C.; Mundim, N.C.; Valente, A.P.; Bemquerer, M.P.; Pilo-Veloso, D.; Bechinger, B. Solution nmr structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations. Peptides 2008, 29, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Epand, R.F.; Mishra, B.; Lushnikova, T.; Thomas, V.C.; Bayles, K.W.; Epand, R.M. Decoding the functional roles of cationic side chains of the major antimicrobial region of human cathelicidin ll-37. Antimicrob. Agents Chemother. 2012, 56, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.C.; Maes, D.; Loris, R.; Pepermans, H.A.; Wyns, L.; Willem, R.; Verheyden, P. H nmr study of the solution structure of ac-amp2, a sugar binding antimicrobial protein isolated from amaranthus caudatus. J. Mol. Boil. 1996, 258, 322–333. [Google Scholar] [CrossRef]
- Oh, D.; Shin, S.Y.; Lee, S.; Kang, J.H.; Kim, S.D.; Ryu, P.D.; Hahm, K.S.; Kim, Y. Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin a(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures. Biochemistry 2000, 39, 11855–11864. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, E.; Shin, S.; Jeong, K.W.; Lee, J.Y.; Bae, S.H.; Kim, S.H.; Lee, J.; Kim, S.R.; Lee, D.G.; et al. Structure and Function of Papiliocin with Antimicrobial and Anti-Inflammatory Activities Isolated from the Swallowtail Butterfly, Papilio Xuthus. Available online: http://www.rcsb.org/structure/2LA2 (accessed on 16 May 2018).
- Santana, M.J.; de Oliveira, A.L.; Queiroz Junior, L.H.; Mandal, S.M.; Matos, C.O.; Dias Rde, O.; Franco, O.L.; Liao, L.M. Structural insights into cn-amp1, a short disulfide-free multifunctional peptide from green coconut water. FEBS Lett. 2015, 589, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Essig, A.; Hofmann, D.; Munch, D.; Gayathri, S.; Kunzler, M.; Kallio, P.T.; Sahl, H.G.; Wider, G.; Schneider, T.; Aebi, M. Copsin, a novel peptide-based fungal antibiotic interfering with the peptidoglycan synthesis. J. Boil. Chem. 2014, 289, 34953–34964. [Google Scholar] [CrossRef] [PubMed]
- Falcao, C.B.; Perez-Peinado, C.; de la Torre, B.G.; Mayol, X.; Zamora-Carreras, H.; Jimenez, M.A.; Radis-Baptista, G.; Andreu, D. Structural dissection of crotalicidin, a rattlesnake venom cathelicidin, retrieves a fragment with antimicrobial and antitumor activity. J. Med. Chem. 2015, 58, 8553–8563. [Google Scholar] [CrossRef] [PubMed]
- Fontana, R.; Mendes, M.A.; de Souza, B.M.; Konno, K.; Cesar, L.M.; Malaspina, O.; Palma, M.S. Jelleines: A family of antimicrobial peptides from the royal jelly of honeybees (apis mellifera). Peptides 2004, 25, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.M.; Dey, S.; Mandal, M.; Sarkar, S.; Maria-Neto, S.; Franco, O.L. Identification and structural insights of three novel antimicrobial peptides isolated from green coconut water. Peptides 2009, 30, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Sung, W.S.; Park, s.H.; Lee, D.G. Antimicrobial effect and membrane-active mechanism of urecjistachykinins, neurpeptides derived from urechis unicinctus. FEBS Lett. 2008, 582, 2463–2466. [Google Scholar] [CrossRef] [PubMed]
- Hou, F.; Li, J.; Pan, P.; Xu, J.; Liu, L.; Liu, W.; Song, B.; Li, N.; Wan, J.; Gao, H. Isolation and characterization of a new antimicrobial peptide from the skin of xenopus laevis. Int. J. Antimicrob. Agents 2011, 36, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Y.; Wang, A.; Song, Y.; Ma, D.; Yang, H.; Ma, Y.; Lai, R. Five novel antimicrobial peptides from skin secretions of the frog, amolops loloensis. Comp. Biochem. Physiol. Part B Biochem. Mol. Boil. 2010, 155, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Chen, X.; Ma, C.; Zhou, M.; Xi, X.; Wang, L.; Ding, A.; Duan, J.; Chen, T.; Shaw, C. Balteatide: A novel antimicrobial decapeptide from the skin secretion of the purple-sided leaf frog, phyllomedusa baltea. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, S.J.; Won, H.S.; Choi, W.S.; Lee, B.J. De novo generation of antimicrobial lk peptides with a single tryptophan at the critical amphipathic interface. J. Pept. Sci. 2009, 15, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Sai, K.P.; Jagannadham, M.V.; Vairamani, M.; Raju, N.P.; Devi, A.S.; Nagaraj, R.; Sitaram, N. Tigerinins: Novel antimicrobial peptides from the indian frog rana tigerina. J. Boil. Chem. 2001, 276, 2701–2707. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Y.; Lee, W.H.; Zhang, Y. Antimicrobial peptides from the venom gland of the social wasp vespa tropica. Toxicon 2013, 74, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.A.; de Souza, B.M.; Marques, M.R.; Palma, M.S. Structural and biological characterization of two novel peptides from the veom of the neotropical social wasp agelaia pallipes pallipes. Toxicon 2004, 44, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, X.; Xu, C.; Zhou, W.; Zhang, K.; Yu, H.; Zhang, Y.; Zheng, Y.; Rees, H.H.; Lai, R.; et al. Anti-infection peptidomics of amphibian skin. Mol. Cell. Proteom. 2007, 6, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Ross, N.W.; MacKinnon, S.L. Myxinidin, a novel antimicrobial peptide from the epidermal mucus of hagfish, myxine glutionsa L. Mar. Biotechnol. 2009, 11, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Asoodeh, A.; Zardini, H.Z.; Chamani, J. Identification and characterization of two novel antimicrobial peptides, temporin-ra and temporin-rb, from skin secretions of the marsh frog (rana ridibunda). J. Pept. Sci. 2012, 18, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Monincová, L.; Buděšínský, M.; Slaninová, J.; Hovorka, O.; Cvačka, J.; Voburka, Z.; Fučík, V.; Borovičková, L.; Bednárová, L.; Straka, J.; et al. Novel antimicrobial peptides from the venom of the eusocial bee halictus sexcinctus (hymenoptera: Halictidae) and their analogs. Amino Acids 2010, 39, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Alkotaini, B.; Anuar, N.; Kadhum, A.A.H.; Sani, A.A.A. Detection of secreted antimicrobial peptides isolated from cell-free culture supernatant of paenibacillus alvei an5. J. Ind. Microbiol. Biotechnol. 2013, 40, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Simmaco, M.; Mignogna, G.; Canofeni, S.; Miele, R.; Mangoni, M.L.; Barra, D. Temporins, antimicrobial peptides from the european red frog rana temporaria. Eur. J. Biochem. 1996, 242, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumari, V.; Nagaraj, R. Antimicrobial and hemolytic activities of crabrolin, a 13-residue peptide from the venome of the european hornet, vespa crabro, and its analogues. J. Pept. Res. 1997, 50, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Falla, T.J.; Hancock, R.E. Improved activity of a synthetic indolicidin analog. Antimicrob. Agents Chemiother. 1997, 41, 1997. [Google Scholar]
- Kim, J.B.; Conlon, J.M.; Iwamuro, S.; Knoop, F.C. Antimicrobial peptides from the skin of the japanese mountain brown frog, rana ornativentris. J. Pept. Res. 2001, 58, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Sonnevend, A.; Davidson, C.; Demandt, A.; Jouenne, T. Host-defense peptides isolated from the skin secretions of the northern red-legged frog rana aurora aurora. Dev. Comp. Immunol. 2005, 29, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Abraham, B.; Sonnevend, A.; Jouenne, T.; Cosette, P.; Leprince, J.; Vaudry, H.; Bevier, C.R. Purification and characterization of antimicrobial peptides from the skin secretions of the carpenter frog rana virgatipes (ranidae, aquarana). Regul. Pept. 2005, 131, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.C.; Zhou, L.; Shi, W.; Luo, X.; Zhang, L.; Nie, Y.; Wang, J.; Shifen, W.; Cao, B.; Cao, H. Three new antimicrobial peptides from the scorpion pandinus imperator. Peptides 2013, 45, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Abbassi, F.; Oury, B.; Blasco, T.; Sereno, D.; Bolbach, G.; Nicolas, P.; Hani, K.; Amiche, M.; Ladram, A. Isolation, characterization and molecular cloning of new temporins from the skin of the north african ranid pelophylax saharica. Peptides 2008, 29, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Sherman, P.; Luo, L.; Bowie, J.; Zhu, S. Structural and functional characterization of two genetically related meucin peptides highlights evolutionary divergence and convergence in antimicrobial peptides. FASEB J. 2009, 23, 1230–1245. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.C.; Wang, S.X.; Zhu, Y.; Zhu, S.Y.; Li, W.X. Identification and functional characterization of novel scorpion venom peptides with no disulfide bridge from buthus martensii karsch. Peptides 2004, 25, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Corzo, G.; Naoki, H.; Andriantsiferana, M.; Nakajima, T. Purification, structure–function analysis, and molecular characterization of novel linear peptides from scorpion opisthacanthus madagascariensis. Biochem. Biophys. Res. Commun. 2002, 293, 2002. [Google Scholar] [CrossRef]
- Ramírez-Carreto, S.; Quintero-Hernández, V.; Jiménez-Vargas, J.M.; Corzo, G.; Possani, L.D.; Becerril, B.; Ortiz, E. Gene cloning and functional characterization of four novel antimicrobial-like peptides from scorpions of the family vaejovidae. Peptides 2012, 34, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ji, S.; Liu, W.; Yu, X.; Meng, Q.; Lai, R. Different expression profiles of bioactive peptides in pelophylax nigromaculatus from distinct regions. Biosci. Biotechnol. Biochem. 2013, 77, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, X.; Liu, X.; Wu, J.; Liu, C.; Gong, W.; Zhao, Z.; Hong, J.; Lin, D.; Wang, Y. Antioxidant peptidomics reveals novel skin antioxidant system. Mol. Cell. Proteom. 2009, 8, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Nie, Y.; Zeng, X.C.; Cao, H.; Zhang, L.; Zhou, L.; Yang, Y.; Luo, X.; Liu, Y. Genomic and functional characterization of three new venom peptides from the scorpion heterometrus spinifer. Peptides 2014, 53, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Duval, E.; Zatylny, C.; Laurencin, M.; Baudy-Floc’h, M.; Henry, J. Kkkkplfglffglf: A cationic peptide designed to exert antibacterial activity. Peptides 2009, 30, 1608–1612. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.L.; Song, S.S.; Li, Q.; Chen, Y.H.; Wang, Q.Y.; Hou, S.T. Identification and characterisation of a novel antimicrobial polypeptide from the skin secretion of a chinese frog (rana chensinensis). Int. J. Antimicrob. Agents 2009, 33, 538–542. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Al-Dhaheri, A.; Al-Mutawa, E.; Al-Kharrage, R.; Ahmed, E.; Kolodziejek, J.; Nowotny, N.; Nielsen, P.F.; Davidson, C. Peptide defenses of the cascades frog rana cascadae: Implications for the evolutionary history of frogs of the amerana species group. Peptides 2007, 28, 1268–1274. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yang, X.; Yang, X.; Zhai, L.; Lu, Z.; Liu, J.; Yu, H. Antimicrobial peptides from the venoms of vespa bicolor fabricus. Peptides 2008, 29, 1887–1892. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Cabrera, M.P.; De Souza, B.M.; Fontana, R.; Konno, K.; Palma, M.S.; De Azevedo, W.F.J.; Neto, J.R. Conformation and lytic activity of eumenine mastoparan: A new antimicrobial peptide from wasp venom. J. Pept. Res. 2004, 64, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Čeřovský, V.; Slaninová, J.; Fučík, V.; Hulačová, H.; Borovičková, L.; Ježek, R.; Bednárová, L. New potent antimicrobial peptides from the venom of polistinae wasps and their analogs. Peptides 2008, 29, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- De Souza, B.M.; da Silva, A.V.R.; Resende, V.M.F.; Arcuri, H.A.; dos Santos Cabrera, M.P.; Neto, J.R.; Palma, M.S. Characterization of two novel polyfunctional mastoparan peptides from the venom of the social wasp polybia paulista. Peptides 2009, 30, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Jiravanichpaisal, P.; Lee, S.Y.; Kim, Y.A.; Andrén, T.; Söderhäll, I. Antibacterial peptides in hemocytes and hematopoietic tissue from freshwater crayfish pacifastacus leniusculus: Characterization and expression pattern. Dev. Comp. Immunol. 2007, 31, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, H.; Yang, H.; Yu, H.; You, D.; Ma, Y.; Ye, H.; Lai, R. Proteomic analysis of skin defensive factors of tree frog hyla simplex. J. Proteome Res. 2011, 10, 4230–4240. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Shim, M.S.; Chung, J.; Lim, D.Y.; Lee, B.J. Purification and characterization of antimicrobial peptides from the skin secretion of rana dybowskii. Peptides 2007, 28, 1532–1539. [Google Scholar] [CrossRef] [PubMed]
- Konno, K.; Hisada, M.; Naoki, H.; Itagaki, Y.; Fontana, R.; Rangel, M.; Oliveira, J.S.; dos Santos Cabrera, M.P.; Neto, J.R.; Hide, I.; et al. Eumenitin, a novel antimicrobial peptide from the venom of the solitary eumenine wasp eumenes rubronotatus. Peptides 2006, 27, 2624–2631. [Google Scholar] [CrossRef] [PubMed]













| PDB ID | Peptide | Sequence | AA |
|---|---|---|---|
| 2MJT | Anoplin [11] | GLLKFIKWLL-NH2 | 11 |
| 1G89 | Indolicidin [12] | ILPWKWPWWPWRR-NH2 | 13 |
| 2F3A | LLAA [13] | RLFDKIRQVIRKF-NH2 | 14 |
| 1D7N | Mastoparan X [14] | INLKALAALAKKIL-NH2 | 15 |
| 2JQ2 | PW2 [15] | HPLKQYWWRPST | 12 |
| 2JMY | CM15 [16] | KWKLFKKIGAVLKVL | 15 |
| 2MAA | Temporin-1 Ta [17] | FLPLIGRVLSGIL (LPS micelles) | 13 |
| 1D6X | Tritrpticin [18] | VRRFPWWWPFLRR | 13 |
| 1T51 | IsCT [19] | ILGKIWEGIKSLF-NH2 | 14 |
| PDB ID | Peptide | Sequence | AA |
|---|---|---|---|
| 2L24 | Not named [20] | IFGAIAGFIKNIW-NH2 | 14 |
| 2N9A | Decoralin-NH2 [21] | SLLSLIRKLIT-NH2 | 12 |
| 2NAL | Retro-KR-12 [22] | RLFDKIRQVIRK | 12 |
| Cluster | Number of AMPs in Cluster | RMSD | Centroid PDB ID |
|---|---|---|---|
| 1 | 24 | 417.506 | 5MMK |
| 2 | 14 | 466.125 | 2MXQ |
| 3 | 13 | 270.961 | 2NDC |
| 4 | 13 | 155.284 | 2RTV |
| 5 | 11 | 183.288 | 5YKL |
| 6 | 8 | 626.605 | 2LG4 |
| 7 | 7 | 144.665 | 2MUH |
| 8 | 7 | 3.293 | 2MLU |
| 9 | 6 | 500.236 | 5UI7 |
| 10 | 5 | 401.027 | 2MFS |
| 11 | 3 | 333.869 | 1FRY |
| 12 | 3 | 666.667 | 1ZRX |
| 13 | 2 | 1.289 | 2LS1 |
| 14 | 1 | 0.000 | 2N0V |
| PDB Entry | Number of AA | RMSD (Å) | ||
|---|---|---|---|---|
| PEP-FOLD | 1.2 ns | 10 ns | ||
| 2MJT | 11 | 0.4461 | 0.1793 | 2.8464 |
| 1G89 | 13 | 6.4019 | 6.3364 | 10.5531 |
| 2F3A | 14 | 1.6773 | 1.4999 | 1.6041 |
| 1D7N | 15 | 2.1988 | 2.2371 | 3.1736 |
| 2JQ2 | 12 | 3.1997 | 3.5588 | 3.5946 |
| 2JMY | 15 | 1.3061 | 1.3681 | 3.0161 |
| 2MAA | 13 | 2.0415 | 2.6618 | 4.0329 |
| 1D6X | 13 | 3.6692 | 3.3713 | 4.0437 |
| 1T51 | 14 | 1.9547 | 2.4822 | 2.9325 |
| 2L24 | 14 | 1.0952 | 3.0831 | 5.0254 |
| 2N9A | 12 | 1.2719 | 2.2212 | 2.6661 |
| 2NAL | 12 | 1.3380 | 1.8398 | 2.4378 |
| Cluster | Centroid | Size | RMSD |
|---|---|---|---|
| 1 | VCP-VT1 | 25 | 321.152 |
| 2 | UyCT1 | 20 | 550.390 |
| 3 | Andreonicin C1 | 3 | 666.667 |
| 4 | Tigerin3 | 2 | 3.302 |
| 5 | CP-11C | 2 | 500.00 |
| 6 | Jelleine II | 2 | 500.00 |
| 7 | Urechistachykinin I | 2 | 0.320 |
| 8 | Peptide AN5-1 | 2 | 0.323 |
| 9 | Tigerin 1 | 1 | 0.000 |
| 10 | Cn-AMP1 | 1 | 0.000 |
| 11 | Astadidin 2 | 1 | 0.000 |
| 12 | PGLa-H | 1 | 0.000 |
| 13 | Odirranain-V1 | 1 | 0.000 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Passarini, I.; Rossiter, S.; Malkinson, J.; Zloh, M. In Silico Structural Evaluation of Short Cationic Antimicrobial Peptides. Pharmaceutics 2018, 10, 72. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics10030072
Passarini I, Rossiter S, Malkinson J, Zloh M. In Silico Structural Evaluation of Short Cationic Antimicrobial Peptides. Pharmaceutics. 2018; 10(3):72. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics10030072
Chicago/Turabian StylePassarini, Ilaria, Sharon Rossiter, John Malkinson, and Mire Zloh. 2018. "In Silico Structural Evaluation of Short Cationic Antimicrobial Peptides" Pharmaceutics 10, no. 3: 72. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics10030072