Titanium Dioxide Nanoparticles Induce Inhibitory Effects against Planktonic Cells and Biofilms of Human Oral Cavity Isolates of Rothia mucilaginosa, Georgenia sp. and Staphylococcus saprophyticus
Abstract
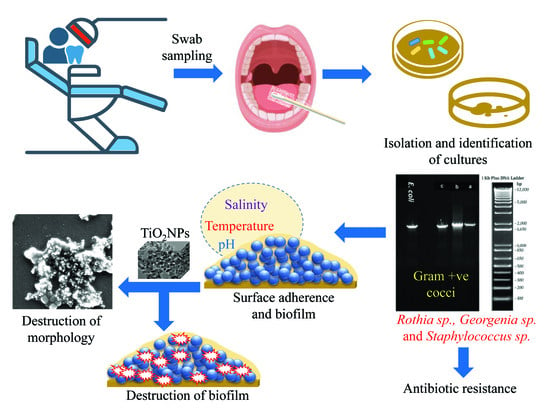
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Isolation and Culture Conditions
2.3. Biochemical Characterization and Antibiotic Sensitivity Profiling of Oral Isolates
2.4. Assessment of Biofilm Formation at Different pH Values, Temperatures, and Salinities
2.5. Phylogenetic Characterization of Oral Bacteria
2.6. Physicochemical Characterization of TiO2NPs
2.7. TiO2NP-Induced Cell Growth and Biofilm Inhibition
2.7.1. Dose-Dependent Anti-Planktonic Cell Activity of TiO2NPs
2.7.2. Dose-Dependent Effect of TiO2NPs on Biofilm Formation
2.8. Statistical Analyses
3. Results and Discussion
3.1. Isolation and Characterization of Oral Bacteria
3.2. Biochemical Characterizations of the Three Oral Isolates
3.3. Phylogenetic Identification
3.4. Effects of Temperature, pH, and NaCl on Biofilm Formation
3.5. Physicochemical Characteristics of TiO2NPs
3.6. Dose-Dependent Effects of TiO2NPs on Biofilm Formation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Palmer, R.J., Jr. Composition and development of oral bacterial communities. Periodontology 2000 2014, 64, 20–39. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-González, I.; Thurnheer, T.; Bartolome, B.; Moreno-Arribas, M.V. Red wine and oenological extracts display antimicrobial effects in an oral bacteria biofilm model. J. Agric. Food Chem. 2014, 62, 4731–4737. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Tawakoli, P.N.; Al-Ahmad, A.; Hoth-Hannig, W.; Hannig, M.; Hannig, C. Comparison of different live/dead stainings for detection and quantification of adherent microorganisms in the initial oral biofilm. Clin. Oral Investig. 2013, 17, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.P.; Mullany, P. Oral biofilms: A reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev. Anti-Infect. Ther. 2010, 8, 1441–1450. [Google Scholar] [CrossRef]
- Potera, C. Antibiotic resistance: Biofilm dispersing agent rejuvenates older antibiotics. Environ. Health Perspect. 2010, 118, A288. [Google Scholar] [CrossRef]
- Da Silva, B.R.; De Freitas, V.A.A.; Carneiro, V.A.; Arruda, F.V.S.; Lorenzón, E.N.; De Aguiar, A.S.W.; Cilli, E.M.; Cavada, B.S.; Teixeira, E.H. Antimicrobial activity of the synthetic peptide Lys-a1 against oral streptococci. Peptides 2013, 42, 78–83. [Google Scholar] [CrossRef]
- Beikler, T.; Flemmig, T.F. Oral biofilm-associated diseases: Trends and implications for quality of life, systemic health and expenditures. Periodontology 2000 2011, 55, 87–103. [Google Scholar] [CrossRef]
- Lu, M.; Ge, Y.; Qiu, J.; Shao, D.; Zhang, Y.; Bai, J.; Zheng, X.; Chang, Z.; Wang, Z.; Dong, W. Redox/pH dual-controlled release of chlorhexidine and silver ions from biodegradable mesoporous silica nanoparticles against oral biofilms. Int. J. Nanomed. 2018, 13, 7697. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Shi, H.; Qi, Y.; Li, J.; Jing, A.; Liu, Q.; Feng, W.; Li, G.; Gao, S. Specific Anti-biofilm Activity of Carbon Quantum Dots by Destroying, P. gingivalis Biofilm Related Genes. Int. J. Nanomed. 2020, 15, 5473. [Google Scholar] [CrossRef]
- Coman, A.N.; Mare, A.; Tanase, C.; Bud, E.; Rusu, A. Silver-deposited nanoparticles on the titanium nanotubes surface as a promising antibacterial material into implants. Metals 2021, 11, 92. [Google Scholar] [CrossRef]
- Tanase, C.; Berta, L.; Mare, A.; Man, A.; Talmaciu, A.I.; Roșca, I.; Mircia, E.; Volf, I.; Popa, V.I. Biosynthesis of silver nanoparticles using aqueous bark extract of Picea abies L. and their antibacterial activity. Eur. J. Wood Wood Prod. 2020, 78, 281–291. [Google Scholar] [CrossRef]
- Zane, A.; Zuo, R.; Villamena, F.A.; Rockenbauer, A.; Foushee, A.M.D.; Flores, K.; Dutta, P.K.; Nagy, A. Biocompatibility and antibacterial activity of nitrogen-doped titanium dioxide nanoparticles for use in dental resin formulations. Int. J. Nanomed. 2016, 11, 6459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.T.; Al-Khedhairy, A.A.; Musarrat, J. ZnO and TiO2 nanoparticles as novel antimicrobial agents for oral hygiene: A review. J. Nanopart. Res. 2015, 17, 1–16. [Google Scholar] [CrossRef]
- Allaker, R.P.; Memarzadeh, K. Nanoparticles and the control of oral infections. Int. J. Antimicrob. Agents 2014, 43, 95–104. [Google Scholar] [CrossRef]
- Yazdi, A.S.; Guarda, G.; Riteau, N.; Drexler, S.K.; Tardivel, A.; Couillin, I.; Tschopp, J. Nanoparticles activate the NLR pyrin domain containing 3 (Nlrp3) inflammasome and cause pulmonary inflammation through release of IL-1α and IL-1β. Proc. Natl. Acad. Sci. USA 2010, 107, 19449–19454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Shaeri, M.; Satar, R.; Ahmed, S.I.; Oves, M.; Ansari, S.A.; Chibber, S. Utilization of Doped Nanoparticles of ZnO and TiO2 As Antimicrobial Agent. Orient. J. Chem. 2019, 35, 1235. [Google Scholar] [CrossRef]
- Priyanka, K.P.; Harikumar, V.S.; Balakrishna, K.M.; Varghese, T. Inhibitory effect of TiO2 NPs on symbiotic arbuscular mycorrhizal fungi in plant roots. IET Nanobiotechnol. 2017, 11, 66–70. [Google Scholar] [CrossRef]
- de Dicastillo, C.L.; Correa, M.G.; Martínez, F.B.; Streitt, C.; Galotto, M.J. Antimicrobial effect of titanium dioxide nanoparticles. In Titanium Dioxide; IntechOpen: London, UK, 2020. [Google Scholar]
- Papaioannou, W.; Gizani, S.; Haffajee, A.D.; Quirynen, M.; Mamai-Homata, E.; Papagiannoulis, L. The microbiota on different oral surfaces in healthy children. Oral Microbiol. Immunol. 2009, 24, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.; Garrity, G.; Jones, D.; Krieg, N.R.; Ludwig, W.; Rainey, F.A.; Schleifer, K.-H.; Whitman, W.B. Bergey’s Manual of Systematic Bacteriology: Volume 3: The Firmicutes; Springer Science & Business Media: New York, NY, USA, 2011; Volume 3, ISBN 0387684891. [Google Scholar]
- Ali, K.; Abul Qais, F.; Dwivedi, S.; Abdel-Salam, E.M.; Ansari, S.M.; Saquib, Q.; Faisal, M.; Al-Khedhairy, A.A.; Al-Shaeri, M.; Musarrat, J. Titanium dioxide nanoparticles preferentially bind in subdomains IB, IIA of HSA and minor groove of DNA. J. Biomol. Struct. Dyn. 2018, 36, 2530–2542. [Google Scholar] [CrossRef] [PubMed]
- Saleem, S.; Ahmed, B.; Khan, M.S.; Al-Shaeri, M.; Musarrat, J. Inhibition of growth and biofilm formation of clinical bacterial isolates by NiO nanoparticles synthesized from Eucalyptus globulus plants. Microb. Pathog. 2017, 111, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, B.; Syed, A.; Ali, K.; Elgorban, A.M.; Khan, A.; Lee, J.; AL-Shwaiman, H.A. Synthesis of gallotannin capped iron oxide nanoparticles and their broad spectrum biological applications. RSC Adv. 2021, 11, 9880–9893. [Google Scholar] [CrossRef]
- Ahmed, B.; Solanki, B.; Zaidi, A.; Khan, M.S.; Musarrat, J. Bacterial toxicity of biomimetic green zinc oxide nanoantibiotic: Insights into ZnONP uptake and nanocolloid-bacteria interface. Toxicol. Res. 2019, 8, 246–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, P.D.; Devine, D.A. How is the development of dental biofilms influenced by the host? J. Clin. Periodontol. 2011, 38, 28–35. [Google Scholar] [CrossRef]
- Holsinger, F.C.; Bui, D.T. Anatomy, function, and evaluation of the salivary glands. In Salivary Gland Disorders; Springer: Berlin/Heidelberg, Germany, 2007; pp. 1–16. [Google Scholar]
- Dawes, C. Estimates, from salivary analyses, of the turnover time of the oral mucosal epithelium in humans and the number of bacteria in an edentulous mouth. Arch. Oral Biol. 2003, 48, 329–336. [Google Scholar] [CrossRef]
- Sarovich, D.S.; Price, E.P.; Von Schulze, A.T.; Cook, J.M.; Mayo, M.; Watson, L.M.; Richardson, L.; Seymour, M.L.; Tuanyok, A.; Engelthaler, D.M. Characterization of ceftazidime resistance mechanisms in clinical isolates of Burkholderia pseudomallei from Australia. PLoS ONE 2012, 7, e30789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higashide, M.; Kuroda, M.; Omura, C.T.N.; Kumano, M.; Ohkawa, S.; Ichimura, S.; Ohta, T. Methicillin-resistant Staphylococcus saprophyticus isolates carrying staphylococcal cassette chromosome mec have emerged in urogenital tract infections. Antimicrob. Agents Chemother. 2008, 52, 2061–2068. [Google Scholar] [CrossRef] [Green Version]
- Matthews, P.; Tomasz, A. Insertional inactivation of the mec gene in a transposon mutant of a methicillin-resistant clinical isolate of Staphylococcus aureus. Antimicrob. Agents Chemother. 1990, 34, 1777–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Sousa, V.S.; da-Silva, A.P.d.S.; Sorenson, L.; Paschoal, R.P.; Rabello, R.F.; Campana, E.H.; Pinheiro, M.S.; dos Santos, L.O.F.; Martins, N.; Botelho, A.C.N. Staphylococcus saprophyticus recovered from humans, food, and recreational waters in Rio de Janeiro, Brazil. Int. J. Microbiol. 2017, 2017, 4287547. [Google Scholar] [CrossRef] [Green Version]
- Kämpfer, P.; Arun, A.B.; Busse, H.-J.; Langer, S.; Young, C.-C.; Chen, W.-M.; Schumann, P.; Syed, A.A.; Rekha, P.D. Georgenia soli sp. nov., isolated from iron-ore-contaminated soil in India. Int. J. Syst. Evol. Microbiol. 2010, 60, 1027–1030. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [Green Version]
- Coton, E.; Desmonts, M.H.; Leroy, S.; Coton, M.; Jamet, E.; Christieans, S.; Donnio, P.Y.; Lebert, I.; Talon, R. Biodiversity of Coagulase-Negative Staphylococci in French cheeses, dry fermented sausages, processing environments and clinical samples. Int. J. Food Microbiol. 2010. [Google Scholar] [CrossRef]
- Basso, A.P.; Martins, P.D.; Nachtigall, G.; Sand, S.V.D.; Moura, T.M.; Frazzon, A.P.G. Antibiotic resistance and enterotoxin genes in Staphylococcus sp. isolates from polluted water in Southern Brazil. An. Acad. Bras. Cienc. 2014, 86, 1813–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uttatree, S.; Charoenpanich, J. Purification and characterization of a harsh conditions-resistant protease from a new strain of Staphylococcus saprophyticus. Agric. Nat. Resour. 2018, 52, 16–23. [Google Scholar] [CrossRef]
- Trivedi, M.K.; Branton, A.; Trivedi, D.; Nayak, G.; Mondal, S.C.; Jana, S. Antimicrobial sensitivity, biochemical characteristics and biotyping of Staphylococcus saprophyticus: An impact of biofield energy treatment. J. Women’s Health Care 2015, 4, 1000271. [Google Scholar]
- Doel, J.J.; Benjamin, N.; Hector, M.P.; Rogers, M.; Allaker, R.P. Evaluation of bacterial nitrate reduction in the human oral cavity. Eur. J. Oral Sci. 2005, 113, 14–19. [Google Scholar] [CrossRef]
- Michels, F.; Colaert, J.; Gheysen, F.; Scheerlinck, T. Late prosthetic joint infection due to Rothia mucilaginosa. Acta Orthop. Belg. 2007, 73, 263. [Google Scholar]
- Dhital, S.; Warren, F.J.; Butterworth, P.J.; Ellis, P.R.; Gidley, M.J. Mechanisms of starch digestion by α-amylase—Structural basis for kinetic properties. Crit. Rev. Food Sci. Nutr. 2017, 57, 875–892. [Google Scholar] [CrossRef]
- Woo, S.-G.; Cui, Y.; Kang, M.-S.; Jin, L.; Kim, K.K.; Lee, S.-T.; Lee, M.; Park, J. Georgenia daeguensis sp. nov., isolated from 4-chlorophenol enrichment culture. Int. J. Syst. Evol. Microbiol. 2012, 62, 1703–1709. [Google Scholar] [CrossRef] [Green Version]
- Fulaz, S.; Devlin, H.; Vitale, S.; Quinn, L.; O’Gara, J.P.; Casey, E. Tailoring nanoparticle-biofilm interactions to increase the efficacy of antimicrobial agents against Staphylococcus aureus. Int. J. Nanomed. 2020, 15, 4779. [Google Scholar] [CrossRef]
- Ameen, F.; Alshehri, W.A.; Nadhari, S. Al Effect of electroactive biofilm formation on acetic acid production in anaerobic sludge driven microbial electrosynthesis. ACS Sustain. Chem. Eng. 2019, 8, 311–318. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.G.; Cho, M.H.; Lee, J. ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol. Res. 2014, 169, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Ćirković, I.; Mijač, V.; Švabić-Vlahović, M. Influence of the incubation temperature, atmosphere and dynamic conditions on biofilm formation by Salmonella spp. Food Microbiol. 2003, 20, 339–343. [Google Scholar] [CrossRef]
- Olson, E.R. Influence of pH on bacterial gene expression. Mol. Microbiol. 1993, 8, 5–14. [Google Scholar] [CrossRef]
- Oliveira, R.; Melo, L.F.; Oliveira, A.; Salgueiro, R. Polysaccharide production and biofilm formation by Pseudomonas fluorescens: Effects of pH and surface material. Colloids Surf. B Biointerfaces 1994, 2, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Rowland, B.M. Bacterial contamination of dental unit waterlines: What is your dentist spraying into your mouth? Clin. Microbiol. Newsl. 2003, 25, 73–77. [Google Scholar] [CrossRef]
- Zhou, L.; Xia, S.; Zhang, Z.; Ye, B.; Xu, X.; Gu, Z.; Wang, X. Effects of pH, Temperature and Salinity on Extracellular Polymeric Substances of Pseudomonas aeruginosa Biofilm with N-(3-Oxooxtanoyl)-L-Homoserine Lactone Addition. J. Water Sustain. 2014, 4, 91. [Google Scholar]
- Cai, T.; Ge, X.; Park, S.Y.; Li, Y. Comparison of Synechocystis sp. PCC6803 and Nannochloropsis salina for lipid production using artificial seawater and nutrients from anaerobic digestion effluent. Bioresour. Technol. 2013, 144, 255–260. [Google Scholar] [CrossRef]
- Møretrø, T.; Hermansen, L.; Holck, A.L.; Sidhu, M.S.; Rudi, K.; Langsrud, S. Biofilm formation and the presence of the intercellular adhesion locus ica among staphylococci from food and food processing environments. Appl. Environ. Microbiol. 2003, 69, 5648–5655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Zou, Y.; Lee, H.; Ahn, J. Effect of NaCl on the biofilm formation by foodborne pathogens. J. Food Sci. 2010, 75, M580–M585. [Google Scholar] [CrossRef]
- Jensen, T.; Kelly, L.; Lazarides, A.; Schatz, G.C. Electrodynamics of noble metal nanoparticles and nanoparticle clusters. J. Clust. Sci. 1999, 10, 295–317. [Google Scholar] [CrossRef]
- Linsebigler, A.L.; Lu, G.; Yates, J.T., Jr. Photocatalysis on TiO2 surfaces: Principles, mechanisms, and selected results. Chem. Rev. 1995, 95, 735–758. [Google Scholar] [CrossRef]
- Battin, T.J.; Kammer, F.V.D.; Weilhartner, A.; Ottofuelling, S.; Hofmann, T. Nanostructured TiO2: Transport behavior and effects on aquatic microbial communities under environmental conditions. Environ. Sci. Technol. 2009, 43, 8098–8104. [Google Scholar] [CrossRef] [PubMed]
- Sodagar, A.; Akhoundi, M.S.A.; Bahador, A.; Jalali, Y.F.; Behzadi, Z.; Elhaminejad, F.; Mirhashemi, A.H. Effect of TiO2 nanoparticles incorporation on antibacterial properties and shear bond strength of dental composite used in Orthodontics. Dent. Press J. Orthod. 2017, 22, 67–74. [Google Scholar]






| Antibiotics | Concentration (µg/disc) | Zone of Inhibition (mm) ± S.D. | ||
|---|---|---|---|---|
| SJM-04 | SJM-38 | SJM-65 | ||
| Nalidixic acid (NA) | 30 | 0 ± 0 (R) | 23 ± 2 (S) | 15 ± 3 (S) |
| Tetracycline (TE) | 10 | 25 ± 5 (S) | 27 ± 3 (S) | 0 ± 0 (R) |
| Norfloxacin (NX) | 10 | 30 ± 3 (S) | 27 ± 4 (S) | 26 ± 4 (S) |
| Cefuroxime (CXM) | 30 | 18 ± 2 (S) | 32 ± 3 (S) | 0 ± 0 (R) |
| Cefotaxime (CTX) | 30 | 26 ± 4 (S) | 30 ± 4 (S) | 31 ± 2 (S) |
| Levofloxacin (LE) | 5 | 32 ± 3 (S) | 28 ± 5 (S) | 29 ± 3 (S) |
| Methicillin (MET) | 5 | 0 ± 0 (R) | 0 ± 0 (R) | 0 ± 0 (R) |
| Ceftazidime (CAZ) | 30 | 0 ± 0 (R) | 0 ± 0 (R) | 0 ± 0 (R) |
| Biochemical Assay | Bacterial Isolates | ||
|---|---|---|---|
| SJM-04 | SJM-38 | SJM-65 | |
| Indole test | _ | _ | _ |
| Methyl red | _ | _ | _ |
| Voges-Proskauer | + | + | _ |
| Citrate | + | + | + |
| Sucrose fermentation | + | + | _ |
| Lactose fermentation | _ | _ | _ |
| Dextrose fermentation | _ | + | + |
| Catalase | + | + | + |
| Oxidase | _ | + | + |
| Nitrate reduction | + | _ | + |
| Starch Hydrolysis | + | + | + |
| Lipid Hydrolysis | _ | + | + |
| Urease | _ | _ | _ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatima, S.; Ali, K.; Ahmed, B.; Al Kheraif, A.A.; Syed, A.; Elgorban, A.M.; Musarrat, J.; Lee, J. Titanium Dioxide Nanoparticles Induce Inhibitory Effects against Planktonic Cells and Biofilms of Human Oral Cavity Isolates of Rothia mucilaginosa, Georgenia sp. and Staphylococcus saprophyticus. Pharmaceutics 2021, 13, 1564. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101564
Fatima S, Ali K, Ahmed B, Al Kheraif AA, Syed A, Elgorban AM, Musarrat J, Lee J. Titanium Dioxide Nanoparticles Induce Inhibitory Effects against Planktonic Cells and Biofilms of Human Oral Cavity Isolates of Rothia mucilaginosa, Georgenia sp. and Staphylococcus saprophyticus. Pharmaceutics. 2021; 13(10):1564. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101564
Chicago/Turabian StyleFatima, Saher, Khursheed Ali, Bilal Ahmed, Abdulaziz A. Al Kheraif, Asad Syed, Abdallah M. Elgorban, Javed Musarrat, and Jintae Lee. 2021. "Titanium Dioxide Nanoparticles Induce Inhibitory Effects against Planktonic Cells and Biofilms of Human Oral Cavity Isolates of Rothia mucilaginosa, Georgenia sp. and Staphylococcus saprophyticus" Pharmaceutics 13, no. 10: 1564. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101564