AS1411 Aptamer Linked to DNA Nanostructures Diverts Its Traffic Inside Cancer Cells and Improves Its Therapeutic Efficacy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Aptamer-Functionalized DNA Nanocages
2.2. Cell Cultures
2.3. DNA Nanocages Stability
2.4. Purification of DNA Nanocages and DNA Blot
2.5. Confocal Analysis
2.6. Cell Viability Assay
2.7. Statistical Analysis
2.8. Computational Methods
2.9. Gaussian Accelerated Molecular Dynamics Simulations
2.10. Trajectory Analysis
3. Results
3.1. Design, Assembly, and In Vitro Stability of AS1411 Functionalized DNA Nanocages
3.2. Intracellular Stability and Targeting Efficiency
3.3. Intracellular Trafficking of Apt-NCs
3.4. Cytotoxic Effect of Apt-NCs
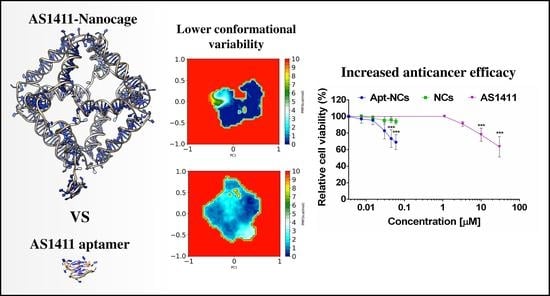
3.5. GaMD Simulations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5’-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef] [Green Version]
- Do, N.Q.; Chung, W.J.; Truong, T.H.A.; Heddi, B.; Phan, A.T. G-quadruplex structure of an anti-proliferative DNA sequence. Nucleic Acids Res. 2017, 45, 7487–7493. [Google Scholar] [CrossRef] [Green Version]
- Bates, P.J.; Laber, D.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Bates, P.J.; Reyes-Reyes, E.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef] [PubMed]
- Dailey, M.M.; Miller, M.C.; Bates, P.J.; Lane, A.N.; Trent, J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010, 38, 4877–4888. [Google Scholar] [CrossRef] [PubMed]
- Christian, S.; Pilch, J.; Akerman, M.E.; Porkka, K.; Laakkonen, P.; Ruoslahti, E. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J. Cell Biol. 2003, 163, 871–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, K.; Gorospe, M. RNA-binding protein nucleolin in disease. RNA Biol. 2012, 9, 799–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Zhao, G.; Zhang, S.; Nigim, F.; Zhou, G.; Yu, Z.; Song, Y.; Chen, Y.; Li, Y. AS1411-Induced Growth Inhibition of Glioma Cells by Up-Regulation of p53 and Down-Regulation of Bcl-2 and Akt1 via Nucleolin. PLoS ONE 2016, 11, e0167094. [Google Scholar] [CrossRef] [Green Version]
- Girvan, A.C.; Teng, Y.; Casson, L.K.; Thomas, S.D.; Jüliger, S.; Ball, M.; Klein, J.B.; Pierce, W.M.; Barve, S.S.; Bates, P.J. AGRO100 inhibits activation of nuclear factor-κB (NF-κB) by forming a complex with NF-κB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006, 5, 1790–1799. [Google Scholar] [CrossRef] [Green Version]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The Nucleolin Targeting Aptamer AS1411 DestabilizesBcl-2Messenger RNA in Human Breast Cancer Cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Lu, J.; Liu, J.; Liang, C.; Wang, M.; Wang, L.; Li, D.; Yao, H.; Zhang, Q.; Wen, J.; et al. A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer. Nat. Commun. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Murphy, E.M.; Centner, C.S.; Bates, P.J.; Malik, M.T.; Kopechek, J.A. Delivery of thymoquinone to cancer cells with as1411-conjugated nanodroplets. PLoS ONE 2020, 15, e0233466. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.; Drabkin, H.A.; Lara, P.N.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Investig. N. Drugs 2013, 32, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.; Iacovelli, F.; Falconi, M.; Cardamone, F.; Della Rocca, B.M.; De Oliveira, C.L.P.; Desideri, A. A Simple and Fast Semiautomatic Procedure for the Atomistic Modeling of Complex DNA Polyhedra. J. Chem. Inf. Model. 2016, 56, 941–949. [Google Scholar] [CrossRef]
- Falconi, M.; Oteri, F.; Chillemi, G.; Andersen, F.F.; Tordrup, D.; Oliveira, C.; Pedersen, J.S.; Knudsen, B.R.; Desideri, A. Deciphering the Structural Properties That Confer Stability to a DNA Nanocage. ACS Nano 2009, 3, 1813–1822. [Google Scholar] [CrossRef] [PubMed]
- Franch, O.; Iacovelli, F.; Falconi, M.; Juul, S.; Ottaviani, A.; Benvenuti, C.; Biocca, S.; Ho, Y.-P.; Knudsen, B.R.; Desideri, A. DNA hairpins promote temperature controlled cargo encapsulation in a truncated octahedral nanocage structure family. Nanoscale 2016, 8, 13333–13341. [Google Scholar] [CrossRef] [PubMed]
- Juul, S.; Iacovelli, F.; Falconi, M.; Kragh, S.L.; Christensen, B.; Frøhlich, R.; Franch, O.; Kristoffersen, E.L.; Stougaard, M.; Leong, K.W.; et al. Temperature-Controlled Encapsulation and Release of an Active Enzyme in the Cavity of a Self-Assembled DNA Nanocage. ACS Nano 2013, 7, 9724–9734. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, C.L.P.; Juul, S.; Jørgensen, H.L.; Knudsen, B.; Tordrup, D.; Oteri, F.; Falconi, M.; Koch, J.; Desideri, A.; Pedersen, J.S.; et al. Structure of Nanoscale Truncated Octahedral DNA Cages: Variation of Single-Stranded Linker Regions and Influence on Assembly Yields. ACS Nano 2010, 4, 1367–1376. [Google Scholar] [CrossRef]
- Vindigni, G.; Raniolo, S.; Ottaviani, A.; Falconi, M.; Franch, O.; Knudsen, B.R.; Desideri, A.; Biocca, S. Receptor-Mediated Entry of Pristine Octahedral DNA Nanocages in Mammalian Cells. ACS Nano 2016, 10, 5971–5979. [Google Scholar] [CrossRef]
- Raniolo, S.; Iacovelli, F.; Unida, V.; Desideri, A.; Biocca, S. In Silico and In Cell Analysis of Openable DNA Nanocages for miRNA Silencing. Int. J. Mol. Sci. 2019, 21, 61. [Google Scholar] [CrossRef] [Green Version]
- Raniolo, S.; Vindigni, G.; Ottaviani, A.; Unida, V.; Iacovelli, F.; Manetto, A.; Figini, M.; Stella, L.; Desideri, A.; Biocca, S. Selective targeting and degradation of doxorubicin-loaded folate-functionalized DNA nanocages. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1181–1190. [Google Scholar] [CrossRef] [Green Version]
- Raniolo, S.; Vindigni, G.; Unida, V.; Ottaviani, A.; Romano, E.; Desideri, A.; Biocca, S. Entry, fate and degradation of DNA nanocages in mammalian cells: A matter of receptors. Nanoscale 2018, 10, 12078–12086. [Google Scholar] [CrossRef]
- Raniolo, S.; Unida, V.; Vindigni, G.; Stolfi, C.; Iacovelli, F.; Desideri, A.; Biocca, S. Combined and selective miR-21 silencing and doxorubicin delivery in cancer cells using tailored DNA nanostructures. Cell Death Dis. 2021, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-J.; Olson, W.K. 3DNA: A versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008, 3, 1213–1227. [Google Scholar] [CrossRef] [PubMed]
- De Lano, W.L. The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, LLC.: San Carlos, CA, USA, 2002. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raniolo, S.; Croce, S.; Thomsen, R.P.; Okholm, A.H.; Unida, V.; Iacovelli, F.; Manetto, A.; Kjems, J.; Desideri, A.; Biocca, S. Cellular uptake of covalent and non-covalent DNA nanostructures with different sizes and geometries. Nanoscale 2019, 11, 10808–10818. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 3, 198–210. [Google Scholar] [CrossRef]
- Ivani, I.; Dans, P.D.; Noy, A.; Pérez, A.; Faustino, I.; Hospital, A.; Walther, J.; Andrio, P.; Goñi, R.; Balaceanu, A.; et al. Parmbsc1: A refined force field for DNA simulations. Nat. Methods 2015, 13, 55–58. [Google Scholar] [CrossRef] [Green Version]
- Havrila, M.; Stadlbauer, P.; Islam, B.; Otyepka, M.; Sponer, J. Effect of Monovalent Ion Parameters on Molecular Dynamics Simulations of G-Quadruplexes. J. Chem. Theory Comput. 2017, 13, 3911–3926. [Google Scholar] [CrossRef]
- Toukmaji, A.Y.; Sagui, C.; Board, J.A.; Darden, T.A. Efficient particle-mesh Ewald based approach to fixed and induced dipolar interactions. J. Chem. Phys. 2000, 113, 10913–10927. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Goga, N.; Rzepiela, A.J.; de Vries, A.H.; Marrink, S.J.; Berendsen, H.J.C. Efficient Algorithms for Langevin and DPD Dynamics. J. Chem. Theory Comput. 2012, 8, 3637–3649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Miao, Y.; Feher, V.A.; McCammon, J.A. Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation. J. Chem. Theory Comput. 2015, 11, 3584–3595. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Bagheri, Z.; Ranjbar, B.; Latifi, H.; Zibaii, M.I.; Moghadam, T.T.; Azizi, A. Spectral properties and thermal stability of AS1411 G-quadruplex. Int. J. Biol. Macromol. 2015, 72, 806–811. [Google Scholar] [CrossRef]
- Jing, Y.; Cai, M.; Zhou, L.; Jiang, J.; Gao, J.; Wang, H. Aptamer AS1411 utilized for super-resolution imaging of nucleolin. Talanta 2020, 217, 121037. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.; Šalipur, F.R.; Shams, M.; Forsthoefel, M.; Bates, P.J. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol. Oncol. 2015, 9, 1392–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Shank, S.; Davis, P.B.; Ziady, A.G. Nucleolin-Mediated Cellular Trafficking of DNA Nanoparticle Is Lipid Raft and Microtubule Dependent and Can Be Modulated by Glucocorticoid. Mol. Ther. 2011, 19, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Charoenphol, P.; Bermudez, H. Aptamer-Targeted DNA Nanostructures for Therapeutic Delivery. Mol. Pharm. 2014, 11, 1721–1725. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Li, X.; Duan, J.; Yang, X.-D. Targeted Treatment of Colon Cancer with Aptamer-Guided Albumin Nanoparticles Loaded with Docetaxel. Int. J. Nanomed. 2020, 15, 6737–6748. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, W.; Mao, C.; Shao, X.-R.; Xie, X.; Wang, F.; Liu, X.; Li, Q.; Lin, Y. DNA-Based Nanomedicine with Targeting and Enhancement of Therapeutic Efficacy of Breast Cancer Cells. ACS Appl. Mater. Interfaces 2019, 11, 15354–15365. [Google Scholar] [CrossRef]
- Xiao, D.; Li, Y.; Tian, T.; Zhang, T.; Shi, S.; Lu, B.; Gao, Y.; Qin, X.; Zhang, M.; Wei, W.; et al. Tetrahedral Framework Nucleic Acids Loaded with Aptamer AS1411 for siRNA Delivery and Gene Silencing in Malignant Melanoma. ACS Appl. Mater. Interfaces 2021, 13, 6109–6118. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Q.; Wang, F.; Zhou, Z.; Xu, J.; Cheng, S.; Cheng, Y. Self-Assembled DNA Nanostructure as a Carrier for Targeted siRNA Delivery in Glioma Cells. Int. J. Nanomed. 2021, 16, 1805–1817. [Google Scholar] [CrossRef] [PubMed]
- Dam, D.H.M.; Culver, K.S.B.; Odom, T.W. Grafting Aptamers onto Gold Nanostars Increases in vitro Efficacy in a Wide Range of Cancer Cell Types. Mol. Pharm. 2014, 11, 580–587. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vindigni, G.; Raniolo, S.; Iacovelli, F.; Unida, V.; Stolfi, C.; Desideri, A.; Biocca, S. AS1411 Aptamer Linked to DNA Nanostructures Diverts Its Traffic Inside Cancer Cells and Improves Its Therapeutic Efficacy. Pharmaceutics 2021, 13, 1671. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101671
Vindigni G, Raniolo S, Iacovelli F, Unida V, Stolfi C, Desideri A, Biocca S. AS1411 Aptamer Linked to DNA Nanostructures Diverts Its Traffic Inside Cancer Cells and Improves Its Therapeutic Efficacy. Pharmaceutics. 2021; 13(10):1671. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101671
Chicago/Turabian StyleVindigni, Giulia, Sofia Raniolo, Federico Iacovelli, Valeria Unida, Carmine Stolfi, Alessandro Desideri, and Silvia Biocca. 2021. "AS1411 Aptamer Linked to DNA Nanostructures Diverts Its Traffic Inside Cancer Cells and Improves Its Therapeutic Efficacy" Pharmaceutics 13, no. 10: 1671. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics13101671