Two-Step Preparation of Protein-Decorated Biohybrid Quantum Dot Nanoparticles for Cellular Uptake
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
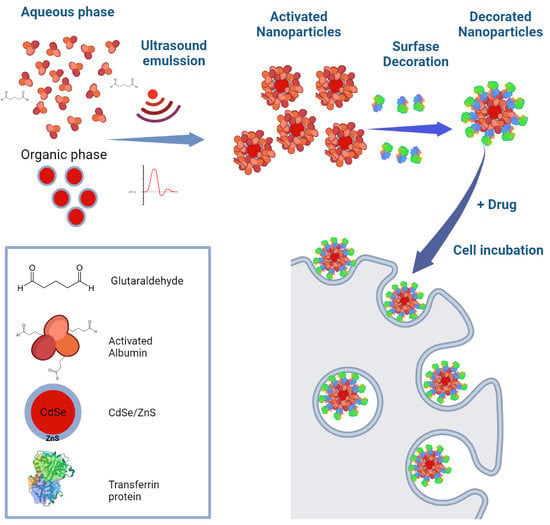
2.2.1. Preparation of Water Soluble Biohybrid QDs
2.2.2. Decoration of Biohybrid HSA-QD NPs
2.3. Characterization of Decorated NPs
2.3.1. Tyndall Effect
2.3.2. Dynamic Light Scattering (DLS)
2.3.3. QDs Fluorescence
2.3.4. Electrophoresis
2.3.5. Transmission Electron Microscopy (TEM)
2.3.6. Confocal Laser Microscopy
2.3.7. Quantification of Decorated NPs in Solution
2.3.8. Quantification of Decorated NPs Uptake in Cell Culture
2.3.9. Preparation of Digitoxin-Loaded hTf-QDs
2.3.10. Cell Lines
2.3.11. In Vivo Analysis of NPs Distributions
3. Results
3.1. Preparation and Characterization of Decorated NPs
3.2. Fluorescence Stability of Decorated NPs
3.3. Cellular Uptake of HSA-QDs and hTf-QDs
3.4. Quantification of Cellular Uptake
3.5. Ability of hTf-QDs to Deliver Cargo into Cells
3.6. In Vitro and In Vivo of NPs Distributions in Ocular Retina
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Q.; Li, S.; He, L.; Feng, X. A brief review of polysialic acid-based drug delivery systems. Int. J. Biol. Macromol. 2023, 230, 123151. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.-S.; Liu, L.; Zeng, R.-F.; Qin, Y.-H.; Hou, J.-W.; Xie, S.-S.; Yue, S.; Yang, J.; Ho, R.J.Y.; Ding, Y.; et al. Tumor-specific carrier-free nanodrugs with GSH depletion and enhanced ROS generation for endogenous synergistic anti-tumor by a chemotherapy-photodynamic therapy. Chem. Eng. J. 2021, 407, 127212. [Google Scholar] [CrossRef]
- Barani, M.; Hajinezhad, M.R.; Zargari, F.; Shahraki, S.; Davodabadi, F.; Mirinejad, S.; Sargazi, S.; Rahdar, A.; Díez-Pascual, A.M. Preparation, characterization, cytotoxicity and pharmacokinetics of niosomes containing gemcitabine: In vitro, in vivo, and simulation studies. J. Drug Deliv. Sci. Technol. 2023, 84, 104505. [Google Scholar] [CrossRef]
- Wang, W.; Liu, H.; Huang, Z.; Fu, F.; Wang, W.; Wu, L.; Huang, Y.; Wu, C.; Pan, X. The effect of organic ligand modification on protein corona formation of nanoscale metal organic frameworks. Chin. Chem. Lett. 2022, 33, 4185–4190. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef]
- Manzari, M.T.; Shamay, Y.; Kiguchi, H.; Rosen, N.; Scaltriti, M.; Heller, D.A. Targeted drug delivery strategies for precision medicines. Nat. Rev. Mater. 2021, 6, 351–370. [Google Scholar] [CrossRef]
- Sengupta, S.; Eavarone, D.; Capila, I.; Zhao, G.; Watson, N.; Kiziltepe, T.; Sasisekharan, R. Temporal targeting of tumour cells and neovasculature with a nanoscale delivery system. Nature 2005, 436, 568–572. [Google Scholar] [CrossRef]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 16014. [Google Scholar] [CrossRef]
- Fang, J.; Islam, W.; Maeda, H. Exploiting the dynamics of the EPR effect and strategies to improve the therapeutic effects of nanomedicines by using EPR effect enhancers. Adv. Drug Deliv. Rev. 2020, 157, 142–160. [Google Scholar] [CrossRef]
- Bahrami, B.; Hojjat-Farsangi, M.; Mohammadi, H.; Anvari, E.; Ghalamfarsa, G.; Yousefi, M.; Jadidi-Niaragh, F. Nanoparticles and targeted drug delivery in cancer therapy. Immunol. Lett. 2017, 190, 64–83. [Google Scholar] [CrossRef] [PubMed]
- Fam, S.Y.; Chee, C.F.; Yong, C.Y.; Ho, K.L.; Mariatulqabtiah, A.R.; Tan, W.S. Stealth Coating of Nanoparticles in Drug-Delivery Systems. Nanomaterials 2020, 10, 787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, T.R.; Bernabeu, E.; Rodríguez, J.A.; Patel, S.; Kozman, M.; Chiappetta, D.A.; Holler, E.; Ljubimova, J.Y.; Helguera, G.; Penichet, M.L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta 2012, 1820, 291–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karami, E.; Behdani, M.; Kazemi-Lomedasht, F. Albumin nanoparticles as nanocarriers for drug delivery: Focusing on antibody and nanobody delivery and albumin-based drugs. J. Drug Deliv. Sci. Technol. 2020, 55, 101471. [Google Scholar] [CrossRef]
- Tarhini, M.; Greige-Gerges, H.; Elaissari, A. Protein-based nanoparticles: From preparation to encapsulation of active molecules. Int. J. Pharm. 2017, 522, 172–197. [Google Scholar] [CrossRef] [PubMed]
- Astete, C.E.; Sabliov, C.M. Synthesis and characterization of PLGA nanoparticles. J. Biomater. Sci. Polym. Ed. 2006, 17, 247–289. [Google Scholar] [CrossRef] [PubMed]
- Ulbrich, K.; Hekmatara, T.; Herbert, E.; Kreuter, J. Transferrin- and transferrin-receptor-antibody-modified nanoparticles enable drug delivery across the blood-brain barrier (BBB). Eur. J. Pharm. Biopharm. Off. J. Arb. Fur. Pharm. Verfahr. E.V. 2009, 71, 251–256. [Google Scholar] [CrossRef]
- Ke, P.C.; Lin, S.; Parak, W.J.; Davis, T.P.; Caruso, F. A Decade of the Protein Corona. ACS Nano 2017, 11, 11773–11776. [Google Scholar] [CrossRef]
- Flores, C.Y.; Achilli, E.; Grasselli, M. Radiation-induced preparation of core/shell gold/albumin nanoparticles. Radiat. Phys. Chem. 2018, 142, 60–64. [Google Scholar] [CrossRef]
- Flores, C.Y.; Luis, J.M.H.; Achilli, E.; Schinca, D.C.; Grasselli, M. Plasmon properties of multilayer albumin/gold hybrid nanoparticles. Mater. Res. Express 2019, 6, 055005. [Google Scholar] [CrossRef] [Green Version]
- Achilli, E.; Casajus, G.; Siri, M.; Flores, C.; Kadłubowski, S.; Alonso, S.d.V.; Grasselli, M. Preparation of protein nanoparticle by dynamic aggregation and ionizing-induced crosslinking. Colloids Surf. A Physicochem. Eng. Asp. 2015, 486, 161–171. [Google Scholar] [CrossRef]
- Achilli, E.; Flores, C.Y.; Temprana, C.F.; Alonso, S.D.V.; Radrizzani, M.; Grasselli, M. Enhanced Gold Nanoparticle-Tumor Cell Recognition by Albumin Multilayer Coating. OpenNano 2022, 6, 100033. [Google Scholar] [CrossRef]
- Achilli, E.; Siri, M.; Flores, C.Y.; Kikot, P.A.; Flor, S.; Martinefski, M.; Lucangioli, S.; Alonso, S.d.V.; Grasselli, M. Radiolysis effect of the high proportion of ethanol in the preparation of albumin nanoparticle. Radiat. Phys. Chem. 2020, 169, 108775. [Google Scholar] [CrossRef]
- Siri, M.; Ruocco, M.J.F.; Achilli, E.; Pizzuto, M.; Delgado, J.F.; Ruysschaert, J.M.; Grasselli, M.; Alonso, S.D.V. Effect of structure in ionised albumin based nanoparticle: Characterisation, Emodin interaction, and in vitro cytotoxicity. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 103, 109813. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, M.; Alonso, S.D.V. Polymer-based nanoparticles: Fabrication to applications-the many faces of DC8,9PC and albumin. Biophys. Rev. 2021, 13, 925–930. [Google Scholar] [CrossRef]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146 Pt 10, 2395–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef]
- Forbes, B.; Shah, A.; Martin, G.P.; Lansley, A.B. The human bronchial epithelial cell line 16HBE14o- as a model system of the airways for studying drug transport. Int. J. Pharm. 2003, 257, 161–167. [Google Scholar] [CrossRef]
- Ferreira, D.G.; Batalha, V.L.; Vicente Miranda, H.; Coelho, J.E.; Gomes, R.; Gonçalves, F.Q.; Real, J.I.; Rino, J.; Albino-Teixeira, A.; Cunha, R.A.; et al. Adenosine A2A Receptors Modulate α-Synuclein Aggregation and Toxicity. Cereb. Cortex 2017, 27, 718–730. [Google Scholar] [CrossRef] [Green Version]
- Torbidoni, V.; Iribarne, M.; Ogawa, L.; Prasanna, G.; Suburo, A.M. Endothelin-1 and endothelin receptors in light-induced retinal degeneration. Exp. Eye Res. 2005, 81, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Booyjzsen, C.; Scarff, C.A.; Moreton, B.; Portman, I.; Scrivens, J.H.; Costantini, G.; Sadler, P.J. Fibrillation of transferrin. Biochim. Biophys. Acta 2012, 1820, 427–436. [Google Scholar] [CrossRef]
- Wetzel, R.; Becker, M.; Behlke, J.; Billwitz, H.; Böhm, S.; Ebert, B.; Hamann, H.; Krumbiegel, J.; Lassmann, G. Temperature behaviour of human serum albumin. Eur. J. Biochem. 1980, 104, 469–478. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Bertrand, N.; Zope, H.; Farokhzad, O.C. Emerging understanding of the protein corona at the nano-bio interfaces. Nano Today 2016, 11, 817–832. [Google Scholar] [CrossRef] [Green Version]
- Asci, R.; Vallefuoco, F.; Andolfo, I.; Bruno, M.; De Falco, L.; Iolascon, A. Trasferrin receptor 2 gene regulation by microRNA 221 in SH-SY5Y cells treated with MPP⁺ as Parkinson’s disease cellular model. Neurosci. Res. 2013, 77, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.B.; Zeissler, M.L.; Chadborn, N.; Gibson, K.; Williams, G.; Zajicek, J.P.; Morrison, K.E.; Hanemann, C.O. Changes in iron-regulatory gene expression occur in human cell culture models of Parkinson’s disease. Neurochem. Int. 2011, 59, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Foster, K.A.; Oster, C.G.; Mayer, M.M.; Avery, M.L.; Audus, K.L. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 1998, 243, 359–366. [Google Scholar] [CrossRef]
- Elbaz, H.A.; Stueckle, T.A.; Wang, H.Y.; O’Doherty, G.A.; Lowry, D.T.; Sargent, L.M.; Wang, L.; Dinu, C.Z.; Rojanasakul, Y. Digitoxin and a synthetic monosaccharide analog inhibit cell viability in lung cancer cells. Toxicol. Appl. Pharmacol. 2012, 258, 51–60. [Google Scholar] [CrossRef] [Green Version]
- López-Lázaro, M. Digitoxin as an anticancer agent with selectivity for cancer cells: Possible mechanisms involved. Expert Opin. Ther. Targets 2007, 11, 1043–1053. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Chang, H.H.; Lai, Y.H.; Lin, C.H.; Chen, M.H.; Chang, G.C.; Tsai, M.F.; Chen, J.J. Digoxin Suppresses Tumor Malignancy through Inhibiting Multiple Src-Related Signaling Pathways in Non-Small Cell Lung Cancer. PLoS ONE 2015, 10, e0123305. [Google Scholar] [CrossRef]
- Elsadek, B.; Kratz, F. Impact of albumin on drug delivery—New applications on the horizon. J. Control. Release 2012, 157, 4–28. [Google Scholar] [CrossRef]
- Dzagli, M.M.; Canpean, V.; Iosin, M.; Mohou, M.A.; Astilean, S. Study of the interaction between CdSe/ZnS core-shell quantum dots and bovine serum albumin by spectroscopic techniques. J. Photochem. Photobiol. A Chem. 2010, 215, 118–122. [Google Scholar] [CrossRef]
- Xiao, Q.; Huang, S.; Qi, Z.D.; Zhou, B.; He, Z.K.; Liu, Y. Conformation, thermodynamics and stoichiometry of HSA adsorbed to colloidal CdSe/ZnS quantum dots. Biochim. Biophys. Acta 2008, 1784, 1020–1027. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Zhai, S.; Hu, X.; Hu, Y.; Wu, B.; Xing, D. Visible light-induced crosslinking and physiological stabilization of diselenide-rich nanoparticles for redox-responsive drug release and combination chemotherapy. Biomaterials 2017, 121, 41–54. [Google Scholar] [CrossRef]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 2004, 37, 790–802. [Google Scholar] [CrossRef]
- Andersen, J.T.; Dalhus, B.; Cameron, J.; Daba, M.B.; Plumridge, A.; Evans, L.; Brennan, S.O.; Gunnarsen, K.S.; Bjørås, M.; Sleep, D.; et al. Structure-based mutagenesis reveals the albumin-binding site of the neonatal Fc receptor. Nat. Commun. 2012, 3, 610. [Google Scholar] [CrossRef] [Green Version]
- Chaudhury, C.; Mehnaz, S.; Robinson, J.M.; Hayton, W.L.; Pearl, D.K.; Roopenian, D.C.; Anderson, C.L. The major histocompatibility complex-related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J. Exp. Med. 2003, 197, 315–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, N.; Zhang, H.; Wang, M.; Hao, Q.; Sun, H. Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe. Sci. Rep. 2012, 2, 999. [Google Scholar] [CrossRef] [Green Version]
- He, X.M.; Carter, D.C. Atomic structure and chemistry of human serum albumin. Nature 1992, 358, 209–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, V.H.; Meghani, N.M.; Amin, H.H.; Tran, T.T.D.; Tran, P.H.L.; Park, C.; Lee, B.J. Modulation of serum albumin protein corona for exploring cellular behaviors of fattigation-platform nanoparticles. Colloids Surf. B Biointerfaces 2018, 170, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Alberio, T.; Forlani, G.; Lualdi, M.; Tosi, G.; Accolla, R.S.; Fasano, M. Neonatal Fc receptor is involved in the protection of fibrinogen after its intake in peripheral blood mononuclear cells. J. Transl. Med. 2018, 16, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anabousi, S.; Bakowsky, U.; Schneider, M.; Huwer, H.; Lehr, C.M.; Ehrhardt, C. In vitro assessment of transferrin-conjugated liposomes as drug delivery systems for inhalation therapy of lung cancer. Eur. J. Pharm. Sci. 2006, 29, 367–374. [Google Scholar] [CrossRef]
- Tuma, P.; Hubbard, A.L. Transcytosis: Crossing cellular barriers. Physiol. Rev. 2003, 83, 871–932. [Google Scholar] [CrossRef]
- John, T.A.; Vogel, S.M.; Tiruppathi, C.; Malik, A.B.; Minshall, R.D. Quantitative analysis of albumin uptake and transport in the rat microvessel endothelial monolayer. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003, 284, L187–L196. [Google Scholar] [CrossRef] [Green Version]
- van Dam, E.M.; Stoorvogel, W. Dynamin-dependent transferrin receptor recycling by endosome-derived clathrin-coated vesicles. Mol. Biol. Cell 2002, 13, 169–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, T.R.; Delgado, T.; Helguera, G.; Penichet, M.L. The transferrin receptor part II: Targeted delivery of therapeutic agents into cancer cells. Clin. Immunol. 2006, 121, 159–176. [Google Scholar] [CrossRef] [PubMed]
- Thorstensen, K.; Romslo, I. The role of transferrin in the mechanism of cellular iron uptake. Biochem. J. 1990, 271, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Nogueira-Librelotto, D.R.; Codevilla, C.F.; Farooqi, A.; Rolim, C.M. Transferrin-Conjugated Nanocarriers as Active-Targeted Drug Delivery Platforms for Cancer Therapy. Curr. Pharm. Des. 2017, 23, 454–466. [Google Scholar] [CrossRef]
- Nikazar, S.; Sivasankarapillai, V.S.; Rahdar, A.; Gasmi, S.; Anumol, P.S.; Shanavas, M.S. Revisiting the cytotoxicity of quantum dots: An in-depth overview. Biophys. Rev. 2020, 12, 703–718. [Google Scholar] [CrossRef]
- Kim, K.E.; Jang, I.; Moon, H.; Kim, Y.J.; Jeoung, J.W.; Park, K.H.; Kim, H. Neuroprotective Effects of Human Serum Albumin Nanoparticles Loaded with Brimonidine on Retinal Ganglion Cells in Optic Nerve Crush Model. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5641–5649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzameret, A.; Ketter-Katz, H.; Edelshtain, V.; Sher, I.; Corem-Salkmon, E.; Levy, I.; Last, D.; Guez, D.; Mardor, Y.; Margel, S.; et al. In vivo MRI assessment of bioactive magnetic iron oxide/human serum albumin nanoparticle delivery into the posterior segment of the eye in a rat model of retinal degeneration. J. Nanobiotechnol. 2019, 17, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghava, S.; Hammond, M.; Kompella, U.B. Periocular routes for retinal drug delivery. Expert Opin. Drug Deliv. 2004, 1, 99–114. [Google Scholar] [CrossRef]
- Bejjani, R.A.; BenEzra, D.; Cohen, H.; Rieger, J.; Andrieu, C.; Jeanny, J.C.; Gollomb, G.; Behar-Cohen, F.F. Nanoparticles for gene delivery to retinal pigment epithelial cells. Mol. Vis. 2005, 11, 124–132. [Google Scholar] [PubMed]
- Bourges, J.L.; Gautier, S.E.; Delie, F.; Bejjani, R.A.; Jeanny, J.C.; Gurny, R.; BenEzra, D.; Behar-Cohen, F.F. Ocular drug delivery targeting the retina and retinal pigment epithelium using polylactide nanoparticles. Investig. Ophthalmol. Vis. Sci. 2003, 44, 3562–3569. [Google Scholar] [CrossRef]
- Normand, N.; Valamanesh, F.; Savoldelli, M.; Mascarelli, F.; BenEzra, D.; Courtois, Y.; Behar-Cohen, F. VP22 light controlled delivery of oligonucleotides to ocular cells in vitro and in vivo. Mol. Vis. 2005, 11, 184–191. [Google Scholar] [PubMed]
- Tawfik, M.; Chen, F.; Goldberg, J.L.; Sabel, B.A. Nanomedicine and drug delivery to the retina: Current status and implications for gene therapy. Naunyn Schmiedebergs Arch Pharm. 2022, 395, 1477–1507. [Google Scholar] [CrossRef] [PubMed]












Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Traverso, A.N.; Fragale, D.J.; Viale, D.L.; Garate, O.; Torres, P.; Valverde, G.; Berra, A.; Torbidoni, A.V.; Yakisich, J.S.; Grasselli, M.; et al. Two-Step Preparation of Protein-Decorated Biohybrid Quantum Dot Nanoparticles for Cellular Uptake. Pharmaceutics 2023, 15, 1651. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics15061651
Traverso AN, Fragale DJ, Viale DL, Garate O, Torres P, Valverde G, Berra A, Torbidoni AV, Yakisich JS, Grasselli M, et al. Two-Step Preparation of Protein-Decorated Biohybrid Quantum Dot Nanoparticles for Cellular Uptake. Pharmaceutics. 2023; 15(6):1651. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics15061651
Chicago/Turabian StyleTraverso, Agata Noelia, David José Fragale, Diego Luis Viale, Octavio Garate, Pablo Torres, Gastón Valverde, Alejandro Berra, Ana Vanesa Torbidoni, Juan Sebastián Yakisich, Mariano Grasselli, and et al. 2023. "Two-Step Preparation of Protein-Decorated Biohybrid Quantum Dot Nanoparticles for Cellular Uptake" Pharmaceutics 15, no. 6: 1651. https://0-doi-org.brum.beds.ac.uk/10.3390/pharmaceutics15061651