Facile Bioinspired Preparation of Fluorinase@Fluoridated Hydroxyapatite Nanoflowers for the Biosynthesis of 5′-Fluorodeoxy Adenosine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Medium, and Chemicals
2.2. Gene Expression and Enzymes Protein Preparation
2.3. Co-Expression of flA and Chaperone Genes in E. coli
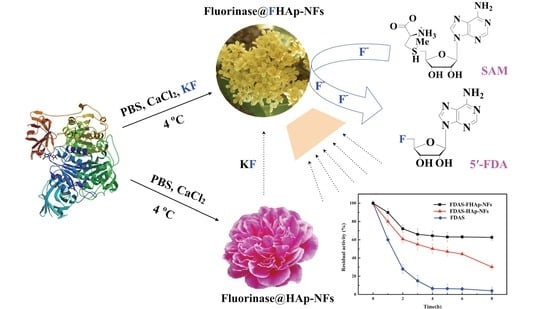
2.4. Preparation and Characterization of Enzyme@fluoridated Hydroxyapatite Nanoflowers (FHAp-NFs)
2.5. Activity Assay and General Procedure of Enzymatic Synthesis of 5′-Fluorodeoxy Adenosine (5′-FDA)
2.6. Optimum Temperature, pH, and Reaction Time of Enzyme Preparations
2.7. Thermal Stability of Fluorinase Preparations
3. Results and Discussion
3.1. Expression and Purification of the Recombinant Fluorinase
3.2. Preparation and Characterization of Enzyme@FHAp-NFs
3.3. Optimum Temperature and pH of Free FDAS and FDAS@ FHAp-NFs Activity
3.4. Thermal Stability of Free FDAS and FDAS@FHAP
3.5. Synthesis of 5′-Fluorodeoxy Adenosine using FDAS Enzyme Preparations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Sánchez-Roselló, M.; Aceña, J.L.; del Pozo, C.; Sorochinsky, A.E.; Fustero, S.; Soloshonok, V.A.; Liu, H. Fluorine in pharmaceutical industry: Fluorine-containing drugs introduced to the market in the last decade (2001–2011). Chem. Rev. 2013, 114, 2432–2506. [Google Scholar] [CrossRef] [PubMed]
- Purser, S.; Moore, P.R.; Swallow, S.; Gouverneur, V. Fluorine in medicinal chemistry. Chem. Soc. Rev. 2008, 37, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Furuya, T.; Kamlet, A.S.; Ritter, T. Catalysis for fluorination and trifluoromethylation. Nature 2011, 473, 470–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Champagne, P.A.; Desroches, J.; Hamel, J.-D.; Vandamme, M.; Paquin, J.-F. Monofluorination of Organic Compounds: 10 Years of Innovation. Chem. Rev. 2015, 115, 9073–9174. [Google Scholar] [CrossRef] [PubMed]
- Rong, J.; Ni, C.; Hu, J. Metal-Catalyzed Direct Difluoromethylation Reactions. Asian J. Org. Chem. 2017, 6, 139–152. [Google Scholar] [CrossRef]
- Walker, M.C.; Thuronyi, B.W.; Charkoudian, L.K.; Lowry, B.; Khosla, C.; Chang, M.C.Y. Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways. Science 2013, 341, 1089–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Hagan, D.; Deng, H. Enzymatic Fluorination and Biotechnological Developments of the Fluorinase. Chem. Rev. 2015, 115, 634–649. [Google Scholar] [CrossRef]
- Ad, O.; Thuronyi, B.W.; Chang, M.C.Y. Elucidating the mechanism of fluorinated extender unit loading for improved production of fluorine-containing polyketides. Proc. Natl. Acad. Sci. USA 2017, 114, E660–E668. [Google Scholar] [CrossRef] [Green Version]
- Ni, C.; Hu, J. The unique fluorine effects in organic reactions: Recent facts and insights into fluoroalkylations. Chem. Soc. Rev. 2016, 45, 5441–5454. [Google Scholar] [CrossRef] [Green Version]
- Prakash, G.S.; Jog, P.V.; Batamack, P.T.; Olah, G.A. Taming of fluoroform: Direct nucleophilic trifluoromethylation of Si, B, S, and C centers. Science 2012, 338, 1324–1327. [Google Scholar] [CrossRef]
- Honda, K.; Harris, T.V.; Hatanaka, M.; Morokuma, K.; Mikami, K. Computational SN2-Type Mechanism for the Difluoromethylation of Lithium Enolate with Fluoroform through Bimetallic C−F Bond Dual Activation. Chem. A Eur. J. 2016, 22, 8796–8800. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Zhao, Y.; Hu, J. AgF-Mediated Fluorinative Cross-Coupling of Two Olefins: Facile Access to α-CF3 Alkenes and β-CF3 Ketones. Angew. Chem. Int. Ed. 2015, 54, 638–642. [Google Scholar]
- Schaffrath, C.; Cobb, S.L.; O’Hagan, D. Cell-free biosynthesis of fluoroacetate and 4-fluorothreonine in Streptomyces cattleya. Angew. Chem. Int. Ed. 2002, 41, 3913–3915. [Google Scholar] [CrossRef]
- Banerjee, R.; Proshlyakov, Y.; Lipscomb, J.D.; Proshlyakov, D.A. Structure of the key species in the enzymatic oxidation of methane to methanol. Nature 2015, 518, 431–434. [Google Scholar] [CrossRef] [Green Version]
- O’Hagan, D.; Schaffrath, C.; Cobb, S.L.; Hamilton, J.T.G.; Murphy, C.D. Biosynthesis of an organofluorine molecule—A fluorinase enzyme has been discovered that catalyses carbon-fluorine bond formation. Nature 2002, 416, 279. [Google Scholar]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [Green Version]
- Onega, M.; Domarkas, J.; Deng, H.; Schweiger, L.F.; Smith, T.A.; Welch, A.E.; Plisson, C.; Gee, A.D.; O’Hagan, D. An enzymatic route to 5-deoxy-5-[18F] fluoro-d-ribose, a [18 F]-fluorinated sugar for PET imaging. Chem. Commun. 2010, 46, 139–141. [Google Scholar] [CrossRef]
- Thompson, S.; Ónega, M.; Ashworth, S.; Fleming, I.N.; Passchier, J.; O’Hagan, D. A two-step fluorinase enzyme mediated 18 F labelling of an RGD peptide for positron emission tomography. Chem. Commun. 2015, 51, 13542–13545. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Rong, J.; Wang, L.; Vasdev, N.; Zhang, L.; Josephson, L.; Liang, S.H. Chemistry for Positron Emission Tomography: Recent Advances in 11C-, 18F-, 13N-, and 15O-Labeling Reactions. Angew. Chem. Int. Ed. 2019, 58, 2580–2605. [Google Scholar] [CrossRef]
- Valencia, D.; Guillén, M.; Fürst, M.J.; López-Santín, J.; Álvaro, G. An immobilized and highly stabilized self-sufficient monooxygenase as biocatalyst for oxidative biotransformations. J. Chem. Technol. Biotechnol. 2018, 93, 985–993. [Google Scholar] [CrossRef]
- Bernal, C.; Rodríguez, K.; Martínez, R. Integrating enzyme immobilization and protein engineering: An alternative path for the development of novel and improved industrial biocatalysts. Biotechnol. Adv. 2018, 36, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Lei, J.; Zare, R.N. Protein-inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, X.; Jiang, M.; Wang, A.; Yang, L.; Pei, X.; Zhang, P.; Wu, S.G. Efficient promiscuous Knoevenagel condensation catalyzed by papain confined in Cu-3(PO4)(2) nanoflowers. Rsc Adv. 2018, 8, 2357–2364. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Hu, S.; Chen, D.; Zhai, H.; Bao, S.; Lv, T. An Evaluation Method of Green development for Chemical Enterprises. Sustainability 2019, 11, 6491. [Google Scholar] [CrossRef] [Green Version]
- Lei, Z.; Gao, C.; Chen, L.; He, Y.; Ma, W.; Lin, Z. Recent advances in biomolecule immobilization based on self-assembly: Organic-inorganic hybrid nanoflowers and metal-organic frameworks as novel substrates. J. Mater. Chem. B 2018, 6, 1581–1594. [Google Scholar] [CrossRef]
- Cui, J.; Jia, S. Organic-inorganic hybrid nanoflowers: A novel host platform for immobilizing biomolecules. Coord. Chem. Rev. 2017, 352, 249–263. [Google Scholar] [CrossRef]
- Zhang, B.; Li, P.; Zhang, H.; Wang, H.; Li, X.; Tian, L.; Ali, N.; Ali, Z.; Zhang, Q. Preparation of lipase/Zn3(PO4)2 hybrid nanoflower and its catalytic performance as an immobilized enzyme. Chem. Eng. J. 2016, 291, 287–297. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Su, Y.; Ouyang, P.; Ge, J.; Liu, Z. Spatial co-localization of multi-enzymes by inorganic nanocrystal-protein complexes. Chem. Commun. 2014, 50, 12465–12468. [Google Scholar] [CrossRef]
- Ma, J.; Wang, J.; Ai, X.; Zhang, S. Biomimetic self-assembly of apatite hybrid materials: From a single molecular template to bi-/multi-molecular templates. Biotechnol. Adv. 2014, 32, 744–760. [Google Scholar] [CrossRef]
- Dey, A.; Bomans, P.H.H.; Müller, F.A.; Will, J.; Frederik, P.M.; de With, G.; Sommerdijk, N.A.J.M. The role of prenucleation clusters in surface-induced calcium phosphate crystallization. Nat. Mater. 2010, 9, 1010–1014. [Google Scholar] [CrossRef]
- Fan, Y.; Sun, Z.; Moradian-Oldak, J. Controlled remineralization of enamel in the presence of amelogenin and fluoride. Biomaterials 2009, 30, 478–483. [Google Scholar] [CrossRef] [Green Version]
- Aoba, T. The effect of fluoride on apatite structure and growth. Crit. Rev. Oral Biol. Med. 1997, 8, 136–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iijima, M.; Onuma, K. Roles of fluoride on octacalcium phosphate and apatite formation on amorphous calcium phosphate substrate. Cryst. Growth Des. 2018, 18, 2279–2288. [Google Scholar] [CrossRef]
- Wang, A.; Du, F.; Pei, X.; Chen, C.; Wu, S.G.; Zheng, Y. Rational immobilization of lipase by combining the structure analysis and unnatural amino acid insertion. J. Mol. Catal. B Enzym. 2016, 132, 54–60. [Google Scholar] [CrossRef]
- Kadambari, L.; Luana, L.; Louise, S. Bridging the Gaps for a ‘Circular’ Bioeconomy: Selection Criteria, Bio-Based Value Chain and Stakeholder Mapping. Sustainability 2018, 10, 1695–1718. [Google Scholar]
- Kaur, J.; Kumar, A.; Kaur, J. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 2017, 106, 803–822. [Google Scholar] [CrossRef]
- Schaffrath, C.; Deng, H.; O’Hagan, D. Isolation and characterisation of 5′-fluorodeoxyadenosine synthase, a fluorination enzyme from Streptomyces cattleya. FEBS Lett. 2003, 547, 111–114. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.; Huang, F.; Deng, H.; Schaffrath, C.; Spencer, J.B.; O’hagan, D.; Naismith, J.H. Crystal structure and mechanism of a bacterial fluorinating enzyme. Nature 2004, 427, 561. [Google Scholar] [CrossRef]
- Ellis, R.J. Molecular chaperones: Assisting assembly in addition to folding. Trends Biochem. Sci. 2006, 31, 395–401. [Google Scholar] [CrossRef]
- Hartl, F.U.; Hayer-Hartl, M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 2002, 295, 1852–1858. [Google Scholar] [CrossRef] [Green Version]
- Gouy, M.; Gautier, C. Codon usage in bacteria: Correlation with gene expressivity. Nucleic Acids Res. 1982, 10, 7055–7074. [Google Scholar] [CrossRef]
- De Bernardez Clark, E. Refolding of recombinant proteins. Curr. Opin. Biotechnol. 1998, 9, 157–163. [Google Scholar] [CrossRef]
- Fattahian, Y.; Riahi-Madvar, A.; Mirzaee, R.; Torkzadeh-Mahani, M.; Asadikaram, G.; Sargazi, G. Optimization of in vitro refolding conditions of recombinant Lepidium draba peroxidase using design of experiments. Int. J. Biol. Macromol. 2018, 118, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, L.; Wang, A.; Li, H.; Wang, C.; Pei, X.; Zhang, P.; Wu, S.G. Efficient synthesis of the key chiral alcohol intermediate of Crizotinib using dual-enzyme@ CaHPO4 hybrid nanoflowers assembled by mimetic biomineralization. J. Chem. Technol. Biotechnol. 2019, 94, 236–243. [Google Scholar] [CrossRef]
- Jokisaari, J.R.; Wang, C.; Qiao, Q.; Hu, X.; Reed, D.A.; Bleher, R.; Luan, X.; Klie, R.F.; Diekwisch, T.G. Particle-Attachment-Mediated and Matrix/Lattice-Guided Enamel Apatite Crystal Growth. ACS Nano 2019, 13, 3151–3161. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.P.; Hackenhaar, C.R.; Lorenzoni, A.S.G.; Rodrigues, R.C.; Costa, T.M.H.; Ninow, J.L.; Hertz, P.F. Chitosan crosslinked with genipin as support matrix for application in food process: Support characterization and β-d-galactosidase immobilization. Carbohydr. Polym. 2016, 137, 184–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.F.; Robinson, D.A.; Mcewan, A.R.; O’Hagan, D.; Naismith, J.H. Mechanism of enzymatic fluorination in Streptomyces cattleya. J. Am. Chem. Soc. 2007, 129, 14597–14604. [Google Scholar] [CrossRef] [Green Version]











© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; Hu, B.; Wang, A.; Li, H.; Yin, Y.; Mao, T.; Xie, T. Facile Bioinspired Preparation of Fluorinase@Fluoridated Hydroxyapatite Nanoflowers for the Biosynthesis of 5′-Fluorodeoxy Adenosine. Sustainability 2020, 12, 431. https://0-doi-org.brum.beds.ac.uk/10.3390/su12010431
Li N, Hu B, Wang A, Li H, Yin Y, Mao T, Xie T. Facile Bioinspired Preparation of Fluorinase@Fluoridated Hydroxyapatite Nanoflowers for the Biosynthesis of 5′-Fluorodeoxy Adenosine. Sustainability. 2020; 12(1):431. https://0-doi-org.brum.beds.ac.uk/10.3390/su12010431
Chicago/Turabian StyleLi, Ningning, Bingjing Hu, Anming Wang, Huimin Li, Youcheng Yin, Tianyu Mao, and Tian Xie. 2020. "Facile Bioinspired Preparation of Fluorinase@Fluoridated Hydroxyapatite Nanoflowers for the Biosynthesis of 5′-Fluorodeoxy Adenosine" Sustainability 12, no. 1: 431. https://0-doi-org.brum.beds.ac.uk/10.3390/su12010431