1. Introduction
Spectral unmixing aims at estimating the fractional abundances of the different pure materials, so-called endmembers, that are contained within a hyperspectral pixel. The main assumption is that the captured reflectance spectrum of each hyperspectral pixel is a mixture of the reflectances of few endmembers. Based on that, for unmixing, a mixing model can be applied and the error between the true spectrum and the spectrum that is reconstructed by the applied mixing model is minimized [
1,
2].
Typically, spectral unmixing is performed by applying the linear mixing model (LMM). This model is valid only when every incoming ray of light interacts only once with a specific pure material in the pixel before reaching the sensor. Taking into account the physical non-negativity and sum-to-one constraints of the fractional abundances, the reconstruction error between the true spectrum and the linearly mixed spectrum can be minimized using the fully constrained least squares unmixing (FCLSU) procedure [
3]. The LMM has shown very good performance in scenarios where the Earth’s surface contains large flat areas with clearly separated regions containing different endmembers. However, the performance of LMM is not satisfactory for scenarios where the scene has a complex geometrical structure. In these scenarios, the incident ray of light may interact with several pure materials within the pixel before reaching the sensor. This causes the captured reflectance spectra to be highly nonlinear mixtures of the endmember reflectances. To explain these nonlinearities, bilinear models have been proposed [
4]. These models extend the LMM by adding bilinear terms, describing that the incident ray of light interacts with two pure materials within the pixel before reaching the sensor. Other nonlinear models try to extend these towards larger numbers of multiple interactions, e.g., the multilinear mixing model (MLM) [
5] and the p-linear (
p > 2) mixture model (pLMM) [
6,
7,
8]. The most advanced nonlinear mixing models are physically-based radiative transfer models. These models are often employed for modeling intimate mixtures of materials. They represent the medium as a half-space filled with particles with known densities and distributions of physical attributes. The Hapke model is a simplified version of a radiative transfer model, [
4,
9,
10].
The available nonlinear mixing models either oversimplify the complex interactions between light and materials or, by considering different kinds of interactions, tend to be inherently complex and non-invertible. Also, all pixels do not necessarily follow the same particular model. Moreover, the parameters of nonlinear mixing models are generally hard to interpret and link to the actual fractional abundances.
Instead of depending on a specific nonlinear mixing model, a new set of algorithms was developed that performed the unmixing of hyperspectral imagery in a reproducing kernel Hilbert space. In [
11], FCLSU was kernelized by radial basis function kernels (KFCLS). However, no improvement in the unmixing result of intimate mixtures was observed [
12].
Another problem is that when using a mixing model, it is assumed that the endmembers are known, i.e., obtained as pure pixels from the data. However, in real scenarios, the pure pixel assumption may collapse due to the relatively low spatial resolution (e.g., ground sampling distances of 30–60m) of hyperspectral images. Many methods have been proposed to solve this problem. Methods that estimate the endmembers along with the fractional abundances have been proposed, that in particular handle the situations when no pure pixels are present in the data, such as volume-constrained nonnegative matrix factorization (MVCNMF) [
13], minimum volume simplex analysis [
14] and robust collaborative nonnegative matrix factorization [
15]. All these methods assume that endmembers span the corner of a linear simplex, and thus cannot cope with nonlinearities. To tackle nonlinearities, in [
16], nonlinear kernelized NMF was presented. More recently, unmixing based on autoencoders [
17,
18,
19,
20,
21,
22] are employed to estimate endmembers and fractional abundances simultaneously. However, these algorithms are either limited to the linear mixing model or implementation of an existing nonlinear (bilinear) model to the autoencoder framework. Finally, sparse unmixing techniques [
23,
24,
25] estimate the fractional abundances of the mixed pixel by linear combinations of spectra from spectral libraries e.g., collected on the ground by a field spectroradiometer. Again, these methods inherently apply the linear mixing model, and cannot cope with nonlinearities.
Finally, the use of fixed endmember spectra results in significant estimation errors [
26] due to spectral variability caused by spatial and temporal variability and variations in illumination conditions. One of the first attempts to address this issue was made by introducing the multiple endmember spectral mixture analysis (MESMA) technique [
27]. Since then, several spectral unmixing techniques that consider endmember variability have been developed and can be classified into three categories: algorithms based on endmember bundles, computational models, and parametric physics-based models [
28]. All these methods, however, inherently apply the linear mixture model.
Alternatively to the assumption of a particular mixing model, some attempts have been made to learn the complex mixing effects using a completely data-driven approach [
29,
30,
31]. We will refer to such methods as supervised spectral unmixing methods, since they require ground truth training data, in the form of the actual compositions for a number of pixels (i.e., the endmember spectra and the fractional abundances). The main strategy of these techniques is to learn a map from the spectra to the fractional abundances. For example, in [
31], a multi-layer perceptron (MLP) was applied as a multivariate regression technique to learn a map from reflectance spectra to the fractional abundances. Similarly, in [
32], support vector regression (SVR) was used. In [
33], an auto-associative neural network was used to extract low dimensional features from the hyperspectral image. The reduced measurement vector was further used as an input to an MLP scheme for pixel-based fuzzy classification. To tackle the endmember variability, in [
34], the mapping was performed using a multi-task Gaussian process framework.
One major drawback of this mapping strategy is that the estimated fractional abundances do not comply with the physical positivity and sum-to-one constraints. This causes substantial amounts of abundance values to become lower than 0 or larger than 1, leading to a loss of the physical meaning of the estimated parameters [
32]. Moreover, most of the methods either use pure pixels or linearly mixed pixels as training data, leading to performances that cannot outperform the simple LMM [
35]. In order to really validate the potential of these methods, actual ground truth abundances of nonlinearly mixed spectra should be provided.
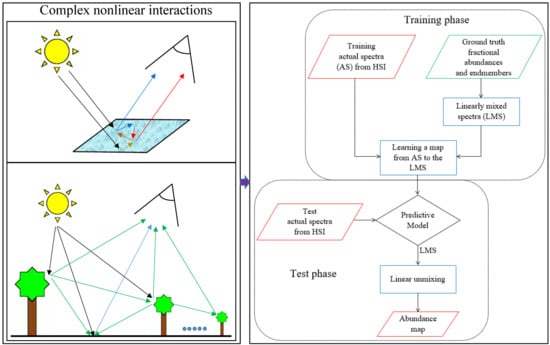
In this work, we propose a strategy to solve these issues. In the proposed method, first linearly mixed spectra are generated from the endmembers and their fractional abundances from the available training data. Then, a map between the actual training spectra and the generated linear spectra is learned by using a regression technique. Finally, after mapping the unknown spectra using the learned regression model, the FCLSU technique is applied to estimate the fractional abundances of the mapped spectra. In order to capture the nonlinearities in the learned maps, the availability of a training set of nonlinearly mixed spectral reflectances and ground-truth information about their endmembers and fractional abundances is a prerequisite.
The proposed unmixing strategy has a number of advantages compared to the state of the art. First of all, it accounts for the nonlinear behavior of spectral reflectances, which is expected to be high in scenes that contain complex geometrical structures or mixtures of minerals. Second, the method avoids dependency on a specific model. Third, the generalization of the mapping procedure allows to learn nonlinearities of different nature simultaneously and makes it robust to noise. Finally, opposite to a direct unconstrained mapping of the spectra to abundances by regression, the approach allows for a straightforward estimation of the fractional abundances with a clear physical meaning. In order to do so, our approach does require the endmember spectra. However, the generalization of the mapping can handle deviations between the applied endmembers and the true ones. As a result, in case that endmember spectra are not available, e.g., when there are no pure pixels present in the data, the endmembers can be collected from a spectral library, e.g., the USGS spectral library.
The mapping to the linear model can be applied by any nonlinear regression technique. In our previous work [
36], this idea was first applied by using an MLP. In this work, we further elaborate on this idea by using different multivariate regression methods for mapping and reporting extensive experimental results. More specifically three mapping methods are presented, based on neural networks (NN), kernel ridge regression (KRR) [
37,
38], and Gaussian processes (GP) [
39]. The presented methodology is validated on an artificial mineral dataset, a synthetic orchard scene, and a real drill core hyperspectral dataset, for which high-quality nonlinear ground reference data is available. The method is compared to some nonlinear mixing models and unconstrained direct mapping onto the abundances.
The remaining of the paper is organized as follows: In
Section 2, we describe the proposed strategy along with the three different mapping approaches. In
Section 3, we describe the datasets on which we validated the proposed method. In
Section 4, we present the experimental results, followed by a discussion in
Section 5.
Section 6 concludes this work.
6. Conclusions
In this paper, we proposed a supervised methodology to estimate fractional abundance maps from hyperspectral images. The method learns a map of the actual spectra to the corresponding linear spectra, composed of the same fractional abundances. Three different mapping procedures, based on artificial neural networks, Gaussian processes, and kernel ridge regression were proposed, but in principle, any nonlinear regression technique can be applied. The methods were validated on a ray-tracing vegetation dataset, a simulated, and a real mineral dataset.
The proposed methodology is superior to unsupervised nonlinear models, which in most cases produced large errors or gave unacceptable results. The methodology also outperforms a direct mapping of the spectra to the fractional abundances. On the vegetation dataset and the simulated mineral dataset, Gaussian processes performed the best. From the experiments on the real drill core dataset, we can conclude that the mapping does not only account for nonlinear effects but also allows the use of library endmembers for the estimation of the fractional abundances. The mapping method based on artificial neural networks provided the best abundance maps of the real drill core sample.
To even better account for endmember variability, the proposed approach can be extended by assigning more than one fixed endmember to each pure material, and learning a mapping to different linear manifolds, after which MESMA can be applied. This will allow us to estimate the endmembers and the corresponding fractional abundances of each pixel. In the future, we will apply the proposed methodology to applications where high-quality ground truth data is more readily available, such as in chemical mixtures and in mixed and compound materials.