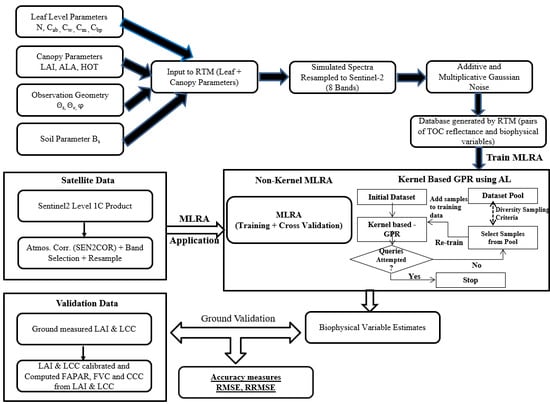
1. Introduction
Accurate quantitative estimation of biophysical variables is of crucial importance for different agricultural and ecological applications. Such variables include leaf area index (LAI), leaf chlorophyll content (LCC), fraction of absorbed photosynthetically active radiation (FAPAR), fractional vegetation cover (FVC), and canopy chlorophyll content (CCC). Their knowledge is valuable, for example, for precision agriculture [
1], crop traits monitoring [
2], and improved yield prediction and reduction of fertilizer usage [
3,
4]. The retrieval of vegetation biophysical variables from multispectral optical satellite data has been performed by many studies in the last decades [
2,
5]. In time, there have been significant developments in the algorithms used for such tasks, with a shift from empirical to more physically-based approaches. Starting from simple parametric regressions between vegetation indices and biophysical variables such as LAI and LCC [
6,
7,
8], the current state of the art relies on hybrid methods exploiting at the same time radiative transfer models (RTM) and non-linear non-parametric regression algorithms [
5,
9,
10]. The methods based on the relationship of, for example, vegetation indices and biophysical variables by the means of fitting functions, typically use the information provided by two or a few spectral bands. This limits the strength of such methods in today’s scenarios where tens or even hundreds of spectral bands are available, respectively, in current super-spectral [
11] or forthcoming hyperspectral [
12,
13] spaceborne sensors.
On the other hand, many of the approaches that have been more recently developed for the retrieval of biophysical variables exploit machine learning regression algorithms (MLRAs). These algorithms have gained wide popularity since they allow the use of more complex models, by requiring only one time-consuming training phase, but allowing their fast application any time thereafter for the retrieval [
14,
15]. Another advantage of these algorithms is the possibility of training them with full spectral information, thus overcoming issues of band selection or transformation, as in the case of parametric regression methods [
16]. These algorithms are adaptive and have the ability to cope with the strong non-linearity inherent in remote sensing data [
17,
18]. Despite their advantages, these algorithms are computationally demanding, so for some MLRAs it is difficult to carry out a training phase with a large number of samples, though there are some that perform well also when trained with small datasets [
19]. Some of the MLRAs are also considered black boxes, e.g., neural networks (NNs) [
11,
20], in which no insight is given about the physical processes linking the spectral reflectance with the biophysical variables. Others, such as partial least squares regression (PLSR) or Gaussian processes regression (GPR), can instead provide some information on what spectral bands are more relevant. In hybrid methods, MLRA can be trained using physically-based modelling approaches that describe the transfer and interaction of radiation inside the canopy based on physical laws, thus providing explicit relations between the biophysical variables and canopy spectral reflectance. When used in the direct mode, biophysical variables are used as input to the physical based RTM, which in turn simulates the top-of-canopy (TOC) spectral reflectance [
9]. Using turbid-medium RTMs it is easy to generate during a short time a large simulated database, with realistic ranges of variation of biophysical variables and their corresponding simulated spectra, within the limits of the assumptions of the RTM on canopy properties. These pairs of biophysical variables and reflectance spectra can be used either in numerical optimization or in look up table (LUT) retrieval methods, by minimizing a cost function expressing the differences between observed (e.g., from satellite data) and RTM model’s simulated reflectance and looking up the corresponding values of the biophysical variables [
21]. For operational applications, due to the pixel-by-pixel calculations, these algorithms are computationally very demanding. These methods are also strongly affected by noise and measurement uncertainties [
11]. As inversion methods of physically-based RTMs, they suffer from the limitation of the ill-posed nature of model inversion [
22,
23], by which different combinations of canopy variables lead to local minima having similar reflectance spectra [
24].
In the hybrid methods of retrieval, the database obtained from RTM simulations can be used to train MLRA which are able to establish complex non-linear, non-parametric models linking the biophysical variables and the spectral reflectance [
9,
11]. Hybrid methods have found widespread application and have reached the operational stage, in particular with the application of NNs [
25,
26,
27] trained with the RTM PROSAIL [
28,
29], such as the algorithm implemented in Sentinel Application Platform (SNAP) biophysical processor tool [
29], developed by the European Space Agency (ESA). Leaf area index retrieval using NNs trained with PROSAIL is also carried out operationally for Cyclopes and Moderate Resolution Imaging Spectrometer (MODIS) products [
30]. However, NN training is a delicate phase and requires the tuning of multiple parameters, which greatly impacts the robustness of the approach [
18]. Thus, in recent years, increasing attention has been paid to alternatives to NNs that are simpler to train and have better potential for retrieval accuracy, such as kernel-based methods [
31]. These methods solve non-linear regression problems by transferring the data to a higher dimensional space by the means of a kernel function [
16]. Kernel-based algorithms have been suggested to offer some advantages in comparison to NNs. Kernel ridge regression (KRR) has proven to be simple for training and to yield competitive accuracy [
18]. Gaussian process regression (GPR) is partially transparent compared to the black box nature of NNs. It allows the use of different kernel functions, ranging from simple to very complex, and it also provides uncertainty estimates with the mean value of prediction [
10,
32]. Gaussian process regression and KRR have been used to estimate successfully LAI and LCC [
10,
33]. Unfortunately, some kernel-based algorithms, such as GPR and KRR, are computationally very expensive if trained on large sets of simulations [
9,
10]. In order to have a general-purpose database, including a wide range of vegetation types, generally a huge number of simulations are performed [
10]. However, not all pairs of the reflectance and corresponding biophysical variables will be relevant. Few attempts have been made by the researchers to optimize the simulations that are generated by the physically-based models [
10,
34]. Active learning (AL) has been proposed as a useful strategy to reduce the size of the RTM-generated database, to make the training of the kernel-based algorithms such as GPR more feasible [
10]. Active learning is a sub-field of machine learning, also called optimal experimental design in statistics [
35]. It initially starts with a small subset of the samples, and then, based on query strategies using either uncertainty or diversity measurement criteria, adds iteratively new samples to the initial training set of samples. In this way, the most informative samples in a dataset are selected, avoiding redundancy [
10]. Active learning techniques have a great potential to optimally sample sets generated by RTM.
Very few studies [
9,
10] have reported the potential of kernel-based methods using AL techniques in relation to the retrieval of biophysical variables such as LAI, LCC, FVC, FAPAR, and CCC. In particular, although the estimation of these variables from hyperspectral data has been demonstrated [
28], the suitability of multispectral satellites has yet to be fully proven, especially for biochemical variables such as LCC. The European Space Agency Sentinel-2 mission has been shown to provide data of a high radiometric quality [
36] and has a higher revisit frequency than what is planned for hyperspectral satellites in the near future. Because of the availability of a larger number of spectral bands than other multispectral satellites, and of the inclusion of the red edge region of the spectrum, Sentinel-2 has a good potential for the estimation of these variables [
9].
This work explores the application of kernel-based GPR using AL for biophysical variables retrieval from Sentinel-2, comparing the potential of this algorithm with other MLRAs and in particular of the version implemented operationally in SNAP.
The main objectives of this study are thus: (1) to compare the performances of different MLRAs, in particular with respect to the algorithm based on NNs implemented in the biophysical processor tool of the ESA SNAP toolbox, by using the same database of PROSAIL simulations [
29]; (2) to assess the accuracy of estimation of the biophysical variables LAI, LCC, FAPAR, CCC, and FVC in the wheat crop, for kernel-based (GPR) and non-kernel-based MLRA hybrid methods of retrieval; (3) to explore the feasibility and potential of the use of AL strategies to optimally sample redundant PROSAIL simulations, to minimize computational time and complexity and allow the use of computationally demanding MLRAs (such as GPR) in hybrid methods.
3. Results
The full simulation dataset generated with PROSAIL was used as a source of subsets for training the different MLAs, as described in the methods section, except for GPR for which a data reduction procedure based on AL was employed. Preliminarily, for GPR we compared three different diversity-based criteria used to perform AL, with respect to the full training with 2500 samples.
Figure 2 shows the comparison of the different methods of sample selection in terms of
R2 and RMSE with respect to the number of iterations, for all the variables, applied to the Maccarese dataset. Only when the performance was improved were the samples added. This explains why fewer samples were added than potentially possible, i.e., less than 50 samples were added for each iteration (e.g., in
Figure 2a the final sample size for CBD was of 1347 at 75 iterations).
The Euclidean distance-based diversity metric EBD surpassed ABD and CBD, achieving higher r2 and lower RMSE values, i.e., higher accuracy and lower error, with a lower number of samples and iterations. The best performing EBD was thus subsequently used in all further applications of AL in the present work. It should be noted that the training of GPR with the full set of 2500 samples, i.e., without AL, provided worse performance than AL in all cases except for FAPAR, despite using a larger dataset for training, revealing some redundancy in the full set.
In the comparison to all the algorithms tested, the cross-validation metrics give an indication of their performance, though the actual accuracy can be more realistically assessed using ground validation results.
Table 4 and
Table 5 report the summary of RMSE estimates for both cross-validation and ground validation for the Maccarese and Shunyi datasets, respectively, for all the algorithms tested.
For Maccarese (
Table 4), LSLR and PLSR were the best performing algorithms for LAI retrieval when validated with ground data, in terms of RMSE, with values of 0.68 (
R2 = 0.78, RRMSE = 19.48%) and 0.69 (
R2 = 0.78, RRMSE = 19.84%) respectively. These were also the fastest computing algorithms among all the MLRAs in terms of time required for training (data not shown). The cross-validation and ground validation results (
Figure 3a) indicated that LSLR apparently did not show saturation, even up to LAI values of 5 or 6. The retrieval of LCC was best performed by RFTB (
Figure 3b) and BagT, with RMSE values of 8.88 μg cm
−2 (
R2 = 0.26 and RRMSE = 17.43%) and 8.90 μg cm
−2 (
R2 = 0.27 and RRMSE = 17.46%). An overestimation of LCC was observed (
Figure 3b). The lowest RMSE of 40.44 g cm
−2 (
R2 = 0.74 RRMSE = 22.7%) (
Figure 3e), was obtained for the retrieval of CCC with PLSR. The retrieval accuracy of LSLR was not significantly different from that of PLSR for CCC (RMSE = 40.86 g cm
−2,
R2 = 0.74 and RRMSE = 22.9%). Indeed, PLSR was not significantly different from LSLR in terms of RMSE for all the variables considered. The lowest RMSE for FVC was 0.08 (
R2 = 0.9 and RRMSE = 9.8%), obtained with the GPR algorithm (
Figure 3c). For FVC, RFTB and BagT were also not significantly different from GPR in terms of RMSE. The FAPAR was best retrieved with a RMSE of 0.10 (
R2 = 0.41 and RRMSE = 12.06%) using the RFTB approach (
Figure 3d), although GPR was not significantly different in terms of RMSE, which was only slightly higher. It was observed that, even in the best performing algorithms, an underestimation of low values of FVC and FAPAR was apparent (
Figure 3c,d). It should also be noted that these latter variables show a smaller range of variation in the measured values as compared to the other biophysical variables. As can be seen, GPR was always the best performing algorithm in the cross-validation results, but this was not confirmed by the ground validation tests, with the exception of FVC and FAPAR estimation.
As can be observed from the horizontal error bars of
Figure 3, considerable variability occurred among ground measurements inside ESUs, despite the visually apparent homogeneity of the crop canopy in the sampled area of each ESU.
For the Shunyi datasets (
Table 5), the ground variability could not be reported for LAI, for which single-point measurements were carried out, but an estimate of the variability for chlorophyll could be made since multiple measurements were available for each point (
Figure 4b).
The best accuracy for LAI retrieval for the Shunyi site was a RMSE of 1.00 m
2 m
−2 (
R2 = 0.73 and RRMSE = 24.12%) obtained with LSLR (
Table 5), i.e., the same algorithm providing the best results also for Maccarese. For LAI, PLSR had a similar performance as that of LSLR, with a RMSE = 1.01 m
2 m
−2 and (
R2 = 0.72 and RRMSE = 24.50%), i.e., also consistently with the results of LAI retrieval for Maccarese. Similarly, for Maccarese, LCC was best retrieved with an RMSE value of 16.77 μg cm
−2, (
R2 = 0.41 and RRMSE = 40.19%) using RFTB (
Table 5). Although RFTB produced the lowest error for retrieving LCC, its retrieval using BagT approach was not significantly different, with RMSE = 17.28 μg cm
−2,
R2 = 0.41 and RRMSE = 41.41%. For CCC retrieval, the lowest RMSE, 56.51 g cm
−2 (
R2 = 0.7, RRMSE = 31.63%), was achieved by the RFTB method. In this case, BagT was not significantly different from RFTB (
R2 = 0.71, RMSE = 56.72 and RRMSE = 31.75%). For this test site, FVC was retrieved with an RMSE of 0.17 (
R2 = 0.73 and RRMSE = 23.65%) with NNs (
Figure 4c). Also, for the Shunyi test site, GPR was always the best performing algorithm in cross-validation but not in ground validation tests.
A summary of the results in terms of RRMSE for all the biophysical variables tested with different MLRAs for both sites is presented in
Figure 5.
It can be seen from
Figure 5, that the k-fold cross-validation (k = 10) provided in some circumstances worse results (higher RRMSE) than the validation with ground data (e.g.,
Figure 5a). This is particularly evident for LAI and CCC for the Maccarese site. Usually, since it was carried out with subset of the same dataset as used for the training of the MLRAs, the cross-validation error was lower than the ground validation error, but this was not the case here, as can be seen from
Figure 5a–e. The different statistical distributions of training and ground validation data might be a possible reason. It appears that the range of variation of the training sets was much larger than that of the ground validation sets, with more extreme values. This could have possibly led to more unreliable retrievals in the cross-validation when extreme values were sampled, which did not happen in the ground validation.
For the Shunyi test site, RRMSE values were generally higher than for the Maccarese (
Figure 5), probably because of the larger differences in the statistical distributions of the ground validation dataset from the training set, as compared for the Maccarese and possibly for the larger error introduced by the sampling protocol.
In general, for LAI, algorithms such as LSLR and PLSR, seemed to outperform more complex MLRAs such as NNs, for example providing better results than those found by applying the biophysical processor implemented in the ESA SNAP toolbox.
For leaf chlorophyll (LCC), much worse results were obtained for Shunyi than for Maccarese, with a clear advantage of RFTB for both sites for ground validation tests. The performance of the NN implementation of SNAP was particularly poor for Shunyi, though it should be noted that this variable was back-calculated from the CCC variable generated by SNAP.
Fractional ground cover (FVC) showed the smallest error among all the biophysical variables tested, alongside FAPAR which was only available for Maccarese. Neural networks provided the best accuracies with the lowest values of relative RMSEs for the latter variable.
Although different algorithms from different families of MLRAs performed well for different biophysical variables, the GPR was considered particularly interesting, as it provided uncertainty estimates with the mean value of prediction (
Figure 3c). However, despite its good cross-validation results, GPR ranked as the top algorithm only for FVC and FAPAR for the Maccarese test site (
Table 4). The AL procedure only selected the most informative samples of the dataset, thus the training of GPR was performed with a smaller set of PROSAIL simulations compared to the other algorithms, showing that it was quite efficient and robust.
The time taken for training the models ranged from 0.003 seconds for LSLR to 47.5 seconds for NNs and a maximum 548.3 seconds for GPR. Although, GPR took longer to train, it should be noted that it performed AL using EBD and used fewer samples to train and achieved high accuracy in comparison to other models.
When considering single ground sampling dates separately, generally the retrieval performances were improved (
Table 6) compared to the bulked data (
Table 4). In some cases, e.g., LAI and CCC, the worst results were obtained at the latest date, due to the fact that it was generally more difficult to estimate higher LAI values.
4. Discussion
There are different methods for the retrieval of biophysical variables from remote sensing data and all the methods have their pros and cons [
16]. Currently, hybrid methods have the capability of combining physical and statistical methods and are considered state of the art. In this paper, different MLRAs (kernel-based and non-kernel based) were trained and applied for the retrieval of the crop biophysical variables LAI, LCC, FAPAR, FVC, and CCC with the same configuration and settings of RTM PROSAIL simulated spectra as those implemented in ESA SNAP biophysical variables retrieval toolbox, with the exception of the observation geometry. With the same of configuration of PROSAIL parameters (
Section 2.1), a total of 41,472 simulated spectra was generated, which turned out to be rather redundant and inefficient for performing the training steps. Subsets of 2500 randomly extracted simulated spectra were used to perform the training of the algorithms and this procedure was repeated ten times with different subsets. This was done in order to allow a comparison of alternative MLRAs to a well-established operational algorithm [
29].
The SNAP algorithm relies on the use of PROSAIL simulations for training NNs, which are the most widely-used tools for operational biophysical variables retrieval [
25,
26,
27,
28]. The downside of the NNs are that they require a relatively long time for training, tuning of parameters is a difficult task, they are black box in nature, and can be unpredictable if training and validation data deviate from each other even slightly [
5]. In this paper, various alternative MLRAs outperformed NNs. For example, LAI retrieval was best performed with LSLR and PLSR, consistently for both tests, although with different accuracies, i.e., RMSE of 0.68 for Maccarese and 1 for Shunyi [
5], compared with different retrieval strategies for LAI and reporting the best performing algorithms for each category. In the case of parametric regression, the best performing algorithm was Tian 3-band formulation (RMSE = 0.615 and
R2 = 0.823), whereas VH-GPR performed best among non-parametric regression algorithms (RMSE = 0.436 and
R2 = 0.902). However, it should be noted that the accuracies found by these authors, somehow better than those of the present study, could be explained by the fact that they used cross-validation in which the training was carried out using the ground dataset, not independent model simulations such as in the present work.
The LCC was best retrieved by the RFTB method for both sites, revealing a general overestimation, but the error of estimation was higher for the Shunyi test site compared to the Maccarese (
Figure 3b and
Figure 4b), particularly for SNAP. Also, the error of CCC estimates was very high for the SNAP tool compared to the other algorithms tested in this work for the Shunyi ground validation (
Figure 5i). It was previously shown by Reference [
64] that Sentinel-2 bands at a 10 m spatial resolution are suitable for estimating LAI, LCC, and CCC. They retrieved LAI (
R2 = 0.809), LCC (
R2 = 0.696) and CCC (
R2 = 0.818) with vegetation indices approach. Multiple vegetation indices were compared for identification of potential vegetation index for the retrieval of LAI, LCC, and CCC in [
65], the best correlation for LCC was with the Meris Terrestrial Chlorophyll Index (MTCI) (
R2 = 0.77) and Sentinel-2 red-edge position index (S2REP) (
R2 = 0.91), for LAI, inverted red-edge chlorophyll index (IRECI) (
R2 = 0.77) and NDI45 (
R2 = 0.62), Normalised Difference Vegetation Index (NDVI) for CCC (
R2 = 0.70). These results are better than those found in the present work, but again, empirical approaches employing the ground datasets were employed by these authors for the calibration of the models.
The FVC variable was more accurately predicted than LAI or LCC, though with slightly higher error for Shunyi. With hierarchical tree-based methods such as BagT, RFTB, and GPR, the error reached was even lower than 10%, which is in line with Global Monitoring for Environment and Security (GMES) goal accuracy [
66] (
Figure 5c). In the case of CCC retrieval, it can be noted that the lowest RRMSE was provided by RFTB and BagT, though also GPR performs similarly to RFTB. The FAPAR variable was only estimated for the Maccarese test site, due to the unavailability of the ground measurements for validation, it was not estimated for the Shunyi test site. The results showed that this variable was best retrieved with the RFTB method, with the lowest RMSE (
Table 3).
The GPR was proved to be an efficient and powerful regressor for the biophysical variables retrieval in previous reports [
18]. A study conducted by Reference [
18], retrieved LAI, Chl, and FVC using different methods, such as NNs, kernel ridge regression (KRR), support vector regression, and GPR for different Sentinel-2 and Sentinel-3 configurations. They found that an overall good performance throughout all Sentinel configurations was provided by GPR. A study carried out by Reference [
10] also found that GPR using AL techniques is an efficient and robust method for the retrieval of LAI and LCC from Sentinel-3 OLCI spectra. However, this was not the case in our study. The GPR method, coupled with AL procedure, provided the best results in terms of computational time and low error only for FVC and FAPAR ground validation tests (
Figure 5c,e,h). On the other hand, GPR generally performed as the best algorithm for the retrieval of all the biophysical variables, only for the cross-validations tests (
Table 4 and
Table 5). Hence, of importance for further analysis to investigate how accurately GPR performs when validated against independent ground data. As Reference [
10] pointed out, GPR is based on non-parametric regression in a Bayesian framework, it provides insights in bands carrying relevant information and also in theoretical uncertainty estimates, thus partially overcoming the black box problem [
5]. These uncertainties are a useful tool for the assessment of upscaling capabilities of biophysical variables from airborne or spaceborne platforms and their respective scales [
32]. A study conducted by [
9] introduced the AL approach with GPR and SVM regressions to deal with the problems of training sample collection for biophysical variables estimation. Their results obtained on simulated MEdium Resolution Imaging Spectrometer (MERIS) and real SeaWiFS Bio-optical Algorithm Mini-Workshop (SeaBAM) datasets were characterized by higher performances in terms of both accuracy and stability with respect to a completely random selection strategy. The present work seems to support these results highlighting the efficiency of the AL procedure, since, when using a smaller set of selected training data, comparable results were obtained than with a random selection of larger size (
Figure 2).