1. Introduction
Hyperspectral imagery (HSI) provides hundreds of narrow and continuous adjacent bands through dense spectral sampling from visible to short-wave infrared regions [
1,
2,
3,
4,
5,
6,
7,
8]. A fine-spectral-resolution HSI provides useful information for classifying different types of ground objects, and it has a variety of applications in many fields such as mineral exploration, environmental monitoring, precision agriculture, and target recognition [
9,
10,
11,
12,
13]. Classification of each pixel in HSI plays a crucial role in these real applications, but complex spectral characteristics within HSI data pose huge challenges to the traditional spectral feature-based HSI classification [
14,
15,
16,
17,
18,
19].
Recent investigations have demonstrated that combining spatial and spectral information is beneficial to the feature extraction and classification of HSI data [
20,
21,
22,
23,
24,
25,
26,
27,
28]. In recent years, many effective spatial-based features have been proposed by concerning structure, shape, texture, geometric, etc. Li et al. [
29] extracted textural features of HSI using LBP operator and then classified them through extreme learning machine (ELM). Mauro et al. [
30] designed an extended multi-attribute profiles (EMAP) algorithm to explore morphological features from HSI, and the extracted features were classified by a random forest classifier. Li et al. [
31] introduced generalized composite kernel machines to explore spatial information through EMAP, then used the multinomial logistic regression for classification. However, in real applications, it is impossible to find a single feature that is suitable for different image scenes due to the variety and irregular distribution of ground objects. The conventional method for addressing this issue is to explore a feature stacking (FS) approach for the combination of different types of features. Li et al. [
32] tried to get combined features by fusing spectral features and EMAP features which improved the classification accuracy of HSI. Song et al. [
33] used LBP operator for extracting textural features, and then stacked spectral features and textural features for classification. However, the feature stacking method commonly poses the problem of the curse-of-dimensionality for the increase in the dimension of stacked features, and thus such methods do not necessarily ensure better performance for HSI classification. Therefore, an urgent challenge in multi-feature classification of HSI data is how to reduce the dimension of spatial and spectral combined features largely with some valuable intrinsic information preserved [
34].
To solve this problem, many DR methods have been proposed to reduce the number of bands and obtain some desired information in HSI [
35,
36,
37,
38]. Principal component analysis (PCA) and Linear Discriminant Analysis (LDA) are two classical DR methods [
39,
40]. However, the two subspace methods cannot analyze the data that lies on or near a submanifold embedded in the original space. Therefore, the graph-based manifold learning methods have attracted wide attention recently [
41]. Such methods include isometric mapping (Isomap), Laplacian eigenmaps (LE), locality preserving projections (LPP), locally linear embedding (LLE), neighborhood preserving embedding (NPE), and local tangent space alignment (LTSA) [
42,
43,
44,
45,
46,
47]. These graph embedding (GE) methods are unsupervised learning methods without using the discriminant information in training samples. Some supervised learning methods were designed to explore the label information of training data to enhance the discriminating ability for classification, such as marginal Fisher analysis (MFA), locality sensitive discriminant analysis (LSDA), coupled discriminant multi-manifold analysis (CDMMA), and local geometric structure Fisher analysis (LGSFA) [
48,
49,
50,
51]. However, the above DR methods only make use of spectral features in HSI, while it is commonly accepted that exploiting multiple features, spectral, texture and shape features, brings significant benefits in terms of improving the classification performance.
To explore DR of multiple features for HSI classification, Fauvel et al. [
52] used PCA to reduce the dimension of EMAP features and stacked them with spectral features to form fused feature vectors. Huo et al. [
53] selected the first three PCs of HSI to extract Gabor textures, then concatenated Gabor textures and spectral features from the same pixel to form the combined feature for classification. However, the above multi-feature-based methods simply stacked the reduced spectral and spatial features together after applying DR on the different types of features, respectively. The embedding features are obtained in different subspaces that cannot ensure global optimization. Furthermore, the direct stacking strategy may lead to unbalanced representation of different features.
To overcome above drawbacks, we propose a novel DR algorithm termed multi-feature manifold discriminant analysis for HSI data. The MFMDA method first exploits the spatial information in HSI by extracting LBP textural features. Then it constructs the intrinsic graphs and penalty graphs of spectral features and LBP features within GE framework, which can effectively discover the manifold structure of spatial features and spectral features. After that, MFMDA learns low-dimensional embedding space from original spectral features as well as LBP features for compacting the intramanifold samples while separating intermanifold samples, which will increase the margins between different manifolds. As a result, the spatial-spectral embedding features possess stronger discriminating ability for HSI classification. Experimental results on three real hyperspectral data sets show that the proposed MFMDA algorithm can significantly improve the classification accuracy compared with some state-of-art DR methods, especially in the case of limited training samples are available.
The remainder of this paper is organized as follows. In
Section 2, we briefly introduce the spectral features, textural features, and the GE framework. The details of our algorithm are introduced in
Section 3.
Section 4 gives experimental results to demonstrate the effectiveness of our algorithm. We give some concluding remarks and suggestions for further work in
Section 5.
3. Proposed Approach
Let us suppose that a hyperspectral data set , where D is the number of bands and N indicates the number of pixels in HSI data. and denote the spectral features and LBP features of X, respectively. The class label of is indicated by , where c is the number of classes. The purpose of DR is to find a low-dimensional embedding space , where d is the embedding dimensionality of extracted features.
3.1. Motivation
Since different types of features represent HSI data from different perspectives, multiple feature fusion will bring benefits to enhance the discrimination capability for classification. The most common way to combine these features is to simply concatenate different types of features together, and then a classifier is employed to classify the stacked features. However, such stack-based methods have witnessed limited performance due to the simple strategy, and they may even perform worse than using a single feature in HSI data. The reasons for this phenomenon are summarized as follows:
Simply stacking spatial and spectral features may yield redundant information, and it remains difficult to achieve an optimal combination for different kinds of features;
The spatial information and spectral information is not equally represented by simply stacking;
The stacked features greatly increase the dimensionality of spatial-spectral combined features, this will make HSI classification fairly challenging for the curse-of-dimensionality problem, especially when only limited training samples are available.
Many DR methods have been explored to reduce the dimension of stacked features. However, different types of features usually lie on different manifolds. Performing dimensionality reduction directly on the simply stacked features cannot reveal the manifold structure of different features in HSI. As a result, the discriminant information contained by different features is not effectively represented, which will restrict their discriminant capability for classification.
To overcome the shortcomings as discussed above, a new DR method called MFMDA is introduced in next section. By exploring the manifold structures of different features, it can effectively extract the spatial-spectral combined features and subsequently improve the classification performance of HSI.
3.2. MFMDA
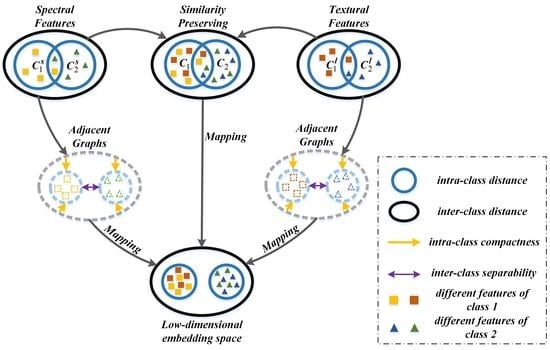
The goal of the proposed MFMDA method is to find an optimized projection matrix which can couple dimensionality reduction and data fusion of original features (from HSI data) and spatial features (LBP features generated from HSI) based on GE framework. MFMDA simultaneously learns a low-dimensional embedding space from original spectral features as well as LBP features for compacting the intramanifold samples while separating intermanifold samples, which will increase the margins between different manifolds. As a result, the obtained embedding features possess stronger discriminating ability that helps to subsequent classification. The flowchart of MFMDA is shown in
Figure 2.
As illustrated in
Figure 2, due to the fact that the similarity relationship between spectral features and LBP features from the same pixel should be preserved in the low-dimensional embedding space. Let us assume
and
are the corresponding projection matrices of spectral features and LBP features, respectively.
and
should be explored to minimize the distance between the two embedding features from the same pixel, and the objective function can be defined as follows:
With some mathematical operations, Equation (
2) can be reduced as:
where
and
are respectively parameterized as
and
,
B and
C are projection matrices that map spectral information and texture information in high-dimensional features to the low-dimensional embedded space, respectively.
,
,
,
I is the identity matrix in
L.
From the view point of classification, in the low-dimensional embedding space, we expect that the samples are as close as possible if they belong to the same manifold, while samples are as far as possible if they are from different manifolds. To achieve this goal, we define the objective function as follows:
where
and
are the affinity weights to characterize the similarity between spectral features
and
of intrinsic graph
as well as the dissimilarity between
and
of the penalty graph
,
and
are the affinity weights to characterize the similarity between LBP features
and
of the intrinsic graph
and the dissimilarity between
and
of the penalty graph
, respectively.
In the intrinsic graph
of spectral features, the vertices
and
are connected by an edge if
and they are close to each other in terms of some distance. When it comes to the penalty graph
, the vertices
and
are connected by an edge if
and
belongs the
nearest neighbors of
. The weights
and
in two spectral-based graphs are defined as:
where
is the
-intramanifold neighbors of spectral feature
,
indicates the
-intermanifold neighbors of
, and
.
In the intrinsic graph
of LBP features, an edge is added between the vertices
and
if
and
belongs the
nearest neighbors of
; in the penalty graph
, an edge is connected by
and
if
and
belongs the
nearest neighbors of
. The weights
and
in two LBP-based graphs can be set as:
where
is the
-intramanifold neighbors of spectral feature
,
indicates the
-intermanifold neighbors of
, and
.
The objective function of
in Equation (
4) is to minimize the intramanifold distance to ensuring the samples from the same manifold should be as close as possible, and the objective function of
in Equation (
5) is to maximize the intermanifold distance for enlarging the manifold margins in the low-dimensional embedding space.
With some mathematical operations, Equations (
4) and (
5) can be reduced as:
where
,
,
,
,
;
,
,
,
,
.
As discussed, the MFMDA method not only preserves the similarity relationship between spectral features and LBP features but also possesses strong discriminating ability in the low-dimensional embedding space. Therefore, a reasonable criterion for choosing a good projection matrix is to optimize the following objective functions:
The multi-objective function optimization problem in Equation (
12) can be equivalent to:
where
,
> 0 are two tradeoff parameters which can adjust intramanifold compactness and intermanifold separability,
.
A constraint
is imposed to remove an arbitrary scaling factor in the projection, and the objective function can be recast as follows:
With the method of Lagrangian multiplier, the optimization solution is formulated as
where
is the Lagrangian multiplier. Then the optimization problem is transformed to solve a generalized eigenvalue problem, i.e.,
where the optimal projection matrix
is composed of
d minimum eigenvalues of Equation (
16) corresponding eigenvectors. Then the low-dimensional feature is given by:
The proposed MFMDA algorithm is summarized in Algorithm 1.
| Algorithm 1 MFMDA. |
- Input:
data set , corresponding class labels , the number of intraclass neighbor points and the number of interclass neighbor points , balance parameter and .
- 1:
Get LBP features generated from the data set, and denote the spectral and LBP features. - 2:
Find the intraclass neighbor points and interclass neighbor points of spectral features and LBP features, respectively. - 3:
Calculate the edge weights of the intrinsic and penalty graphs by Equations ( 6)–( 9). - 4:
Compute the , , and by , , and , respectively. - 5:
Obtain the Lagrangian matrix which contain manifold structure through . - 6:
Calculate matrix E by . - 7:
Construct the matrix according to Equation ( 16). - 8:
Obtain the projection matrix of spectral and LBP features by , and . - 9:
Achieve the low-dimensional features Y through Equation ( 17).
- Output:
; Projection matrix of spectral and LBP features: and .
|
5. Conclusions
Traditional methods explore only a single feature or simply stacked features in hyperspectral image, which will restrict their discriminant capability for classification. In this paper, we proposed a new dimensionality reduction method termed MFMDA to couple DR and fusion of spectral and textual features of HSI data. MFMDA first explores LBP operator to extract textural features for encoding the spatial information in HSI. Then, within GE framework, the intrinsic and penalty graphs of LBP and spectral features are constructed to explore the discriminant manifold structure in both spatial and spectral domains, respectively. After that, a new spatial-spectral DR model is built to extract discriminant spatial-spectral combined features which not only preserve the similarity relationship between spectral features and LBP features but also possess strong discriminating ability in the low-dimensional embedding space. Experiments on Indian Pines, Heihe and PaviaU hyperspectral data sets demonstrate that the proposed MFMDA method can significantly improve classification performance and result in smoother classification maps than some state-of-the-art methods, and with fewer training samples, the classification accuracy can reach 95.43%, 97.19% and 96.60%, respectively. In the future, we will focus on conducting a more detailed investigation of other possible features to further improve the performance of MFMDA.