Machine Learning Techniques for Tree Species Classification Using Co-Registered LiDAR and Hyperspectral Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
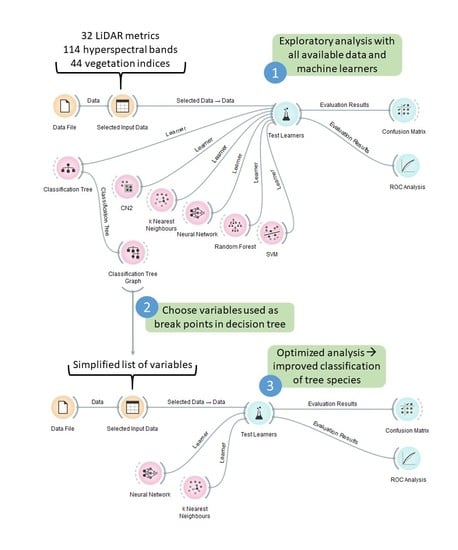
2.2. Data Preparation and Exploration
2.3. Machine Learning Methods, Accuracy, and Validation
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Koch, B. Status and future of laser scanning, synthetic aperture radar and hyperspectral remote sensing data for forest biomass assessment. ISPRS J. Photogramm. Remote Sens. 2010, 65, 581–590. [Google Scholar] [CrossRef]
- Korhonen, L.; Korpela, I.; Heiskanen, J.; Maltamo, M. Airborne discrete-return LIDAR data in the estimation of vertical canopy cover, angular canopy closure and leaf area index. Remote Sens. Environ. 2011, 115, 1065–1080. [Google Scholar] [CrossRef]
- Schardt, M.; Ziegler, M.; Wimmer, A.; Wack, R.; Hyyppä, J. Assessment of forest parameters by means of laser scanning. In Proceedings of the International Archives of Photogrammetry Remote Sensing and Spatial Information Sciences, Graz, Austria, 9–13 September 2002; pp. 302–309. [Google Scholar]
- Wulder, M.A.; Bater, C.W.; Coops, N.C.; Hilker, T.; White, J.C. The role of LiDAR in sustainable forest management. For. Chron. 2008, 84, 807–826. [Google Scholar] [CrossRef] [Green Version]
- Reitberger, J.; Krzystek, P.; Stilla, U. Analysis of full waveform LIDAR data for the classification of deciduous and coniferous trees. Int. J. Remote Sens. 2008, 29, 1407–1431. [Google Scholar] [CrossRef]
- Zhou, T.; Popescu, S.C.; Lawing, A.M.; Eriksson, M.; Strimbu, B.M.; Bürkner, P.C. Bayesian and classical machine learning methods: A Comparison for tree species classification with LiDAR waveform signatures. Remote Sens. 2018, 10, 39. [Google Scholar] [CrossRef]
- Heinzel, J.; Koch, B. Exploring full-waveform LiDAR parameters for tree species classification. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 152–160. [Google Scholar] [CrossRef]
- Cao, L.; Dai, J.; Innes, J.L.; She, G.; Coops, N.C.; Ruan, H. Tree species classification in subtropical forests using small-footprint full-waveform LiDAR data. Int. J. Appl. Earth Obs. Geoinf. 2016, 49, 39–51. [Google Scholar] [CrossRef]
- Lim, K.S.-W.; Treitz, P.; Wulder, M.; St-Onge, B.; Flood, M. Lidar remote sensing of forest structure. Prog. Phys. Geogr. 2003, 27, 88–106. [Google Scholar] [CrossRef]
- Yao, W.; Krzystek, P.; Heurich, M. Tree species classification and estimation of stem volume and DBH based on single tree extraction by exploiting airborne full-waveform LiDAR data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar]
- Sumnall, M.J.; Hill, R.A.; Hinsley, S.A. Comparison of small-footprint discrete return and full waveform airborne lidar data for estimating multiple forest variables. Remote Sens. Environ. 2016, 173, 214–223. [Google Scholar] [CrossRef] [Green Version]
- Hudak, A.T.; Evans, J.S.; Smith, A.M.S. LiDAR utility for natural resource managers. Remote Sens. 2009, 1, 934–951. [Google Scholar] [CrossRef]
- Parkes, B.D.; Newell, G.; Cheal, D. Assessing the quality of native vegetation: The “habitat hectares” approach. Ecol. Manag. Restor. 2003, 4, 29–38. [Google Scholar] [CrossRef]
- Laurin, G.V.; Puletti, N.; Chen, Q.; Corona, P.; Papale, D.; Valentini, R. Above ground biomass and tree species richness estimation with airborne lidar in tropical Ghana forests. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 371–379. [Google Scholar] [CrossRef]
- Fedrigo, M.; Newnham, G.J.; Coops, N.C.; Culvenor, D.S.; Bolton, D.K.; Nitschke, C.R. Predicting temperate forest stand types using only structural profiles from discrete return airborne lidar. ISPRS J. Photogramm. Remote Sens. 2018, 136, 106–119. [Google Scholar] [CrossRef]
- Korpela, I.; Tokola, T.; Orka, H.O.; Koskinen, M. Small-Footprint Discrete-Return Lidar in Tree Species Recognition. In Proceedings of the ISPRS Hannover Workshop 2009, Hannover, Germany, 2–5 June 2009; pp. 1–6. [Google Scholar]
- Van Aardt, J.A.N.; Wynne, R.H.; Scrivani, J.A. Lidar-based Mapping of Forest Volume and Biomass by Taxonomic Group Using Structurally Homogenous Segments. Photogramm. Eng. Remote Sens. 2008, 74, 1033–1044. [Google Scholar] [CrossRef] [Green Version]
- Orka, H.O.; Naesset, E.; Bollandsas, O.M. Utilizing Airborne Laser Intensity for Tree Species Classification. In Proceedings of the SPRS Workshop on Laser Scanning 2007 and SilviLaser 2007, Espoo, Finland, 12–14 September 2007; pp. 1–8. [Google Scholar]
- Gillespie, T.W.; Brock, J.; Wright, C.W. Prospects for quantifying structure, floristic composition and species richness of tropical forests. Int. J. Remote Sens. 2004, 25, 707–715. [Google Scholar] [CrossRef]
- Liu, L.; Pang, Y.; Fan, W.; Li, Z.; Zhang, D.; Li, M. Fused airborne LiDAR and hyperspectral data for tree species identification in a natural temperate forest. J. Remote Sens. 2013, 1007, 679–695. [Google Scholar]
- Ni-Meister, W.; Lee, S.; Strahler, A.H.; Woodcock, C.E.; Schaaf, C.; Yao, T.; Ranson, K.J.; Sun, G.; Blair, J.B. Assessing general relationships between aboveground biomass and vegetation structure parameters for improved carbon estimate from lidar remote sensing. J. Geophys. Res. Biogeosci. 2010, 115, 1–12. [Google Scholar] [CrossRef]
- Jones, T.G.; Coops, N.C.; Sharma, T. Assessing the utility of airborne hyperspectral and LiDAR data for species distribution mapping in the coastal Pacific Northwest, Canada. Remote Sens. Environ. 2010, 114, 2841–2852. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Comparison of area-based and individual tree-based methods for predicting plot-level forest attributes. Remote Sens. 2010, 2, 1481–1495. [Google Scholar] [CrossRef]
- Hovi, A.; Korhonen, L.; Vauhkonen, J.; Korpela, I. LiDAR waveform features for tree species classification and their sensitivity to tree- and acquisition related parameters. Remote Sens. Environ. 2016, 173, 224–237. [Google Scholar] [CrossRef]
- Ferraz, A.; Saatchi, S.; Mallet, C.; Meyer, V. Lidar detection of individual tree size in tropical forests. Remote Sens. Environ. 2016, 183, 318–333. [Google Scholar] [CrossRef]
- Jeronimo, S.M.A.; Kane, V.R.; Churchill, D.J.; McGaughey, R.J.; Franklin, J.F. Applying LiDAR individual tree detection to management of structurally diverse forest landscapes. J. For. 2018, 116, 336–346. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.J.; Zhang, L. Trends in automatic individual tree crown detection and delineation—Evolution of LiDAR data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å.; Söderman, U. Species identification of individual trees by combining high resolution LiDAR data with multi-spectral images. Int. J. Remote Sens. 2008, 29, 1537–1552. [Google Scholar] [CrossRef]
- Cho, M.A.; Mathieu, R.; Asner, G.P.; Naidoo, L.; van Aardt, J.; Ramoelo, A.; Debba, P.; Wessels, K.; Main, R.; Smit, I.P.J.; et al. Mapping tree species composition in South African savannas using an integrated airborne spectral and LiDAR system. Remote Sens. Environ. 2012, 125, 214–226. [Google Scholar] [CrossRef]
- Karna, Y.K.; Hussin, Y.A.; Gilani, H.; Bronsveld, M.C.; Murthy, M.S.R.; Qamer, F.M.; Karky, B.S.; Bhattarai, T.; Aigong, X.; Baniya, C.B. Integration of WorldView-2 and airborne LiDAR data for tree species level carbon stock mapping in Kayar Khola watershed, Nepal. Int. J. Appl. Earth Obs. Geoinf. 2015, 38, 280–291. [Google Scholar] [CrossRef]
- Van Ewijk, K.Y.; Randin, C.F.; Treitz, P.M.; Scott, N.A. Predicting fine-scale tree species abundance patterns using biotic variables derived from LiDAR and high spatial resolution imagery. Remote Sens. Environ. 2014, 150, 120–131. [Google Scholar] [CrossRef]
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef]
- Kukkonen, M.; Maltamo, M.; Korhonen, L.; Packalen, P. Multispectral Airborne LiDAR Data in the Prediction of Boreal Tree Species Composition. IEEE Trans. Geosci. Remote Sens. 2019, 1–10. [Google Scholar] [CrossRef]
- Budei, B.C.; St-Onge, B.; Hopkinson, C.; Audet, F.A. Identifying the genus or species of individual trees using a three-wavelength airborne lidar system. Remote Sens. Environ. 2018, 204, 632–647. [Google Scholar] [CrossRef]
- Amiri, N.; Heurich, M.; Krzystek, P.; Skidmore, A.K. Feature relevance assessment of multispectral airborne LiDAR data for tree species classification. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci.-ISPRS Arch. 2018, 42, 31–34. [Google Scholar] [CrossRef]
- Wessel, M.; Brandmeier, M.; Tiede, D. Evaluation of different machine learning algorithms for scalable classification of tree types and tree species based on Sentinel-2 data. Remote Sens. 2018, 10, 1419. [Google Scholar] [CrossRef]
- Blackburn, G.A. Remote sensing of forest pigments using airborne imaging spectrometer and LIDAR imagery. Remote Sens. Environ. 2002, 82, 311–321. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Liu, L.; Coops, N.C.; Aven, N.W.; Pang, Y. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote Sens. Environ. 2017, 200, 170–182. [Google Scholar] [CrossRef]
- Swatantran, A.; Dubayah, R.; Roberts, D.; Hofton, M.; Blair, J.B. Mapping biomass and stress in the Sierra Nevada using lidar and hyperspectral data fusion. Remote Sens. Environ. 2011, 115, 2917–2930. [Google Scholar] [CrossRef] [Green Version]
- Andrew, M.E.; Ustin, S.L. Habitat suitability modelling of an invasive plant with advanced remote sensing data. Divers. Distrib. 2009, 15, 627–640. [Google Scholar] [CrossRef]
- Koetz, B.; Morsdorf, F.; van der Linden, S.; Curt, T.; Allgöwer, B. Multi-source land cover classification for forest fire management based on imaging spectrometry and LiDAR data. For. Ecol. Manag. 2008, 256, 263–271. [Google Scholar] [CrossRef]
- Mundt, J.T.; Streutker, D.R.; Glenn, N.F. Mapping sagebrush distribution using fusion of hyperspectral and lidar classifications. Photogramm. Eng. Remote Sens. 2006, 72, 47–54. [Google Scholar] [CrossRef]
- Cook, B.D.; Corp, L.A.; Nelson, R.F.; Middleton, E.M.; Morton, D.C.; McCorkel, J.T.; Masek, J.G.; Ranson, K.J.; Ly, V.; Montesano, P.M. NASA Goddard’s LiDAR, hyperspectral and thermal (G-LiHT) airborne imager. Remote Sens. 2013, 5, 4045–4066. [Google Scholar] [CrossRef]
- Morsdorf, F.; Mårell, A.; Koetz, B.; Cassagne, N.; Pimont, F.; Rigolot, E.; Allgöwer, B. Discrimination of vegetation strata in a multi-layered Mediterranean forest ecosystem using height and intensity information derived from airborne laser scanning. Remote Sens. Environ. 2010, 114, 1403–1415. [Google Scholar] [CrossRef] [Green Version]
- Dalponte, M.; Bruzzone, L.; Vescovo, L.; Gianelle, D. The role of spectral resolution and classifier complexity in the analysis of hyperspectral images of forest areas. Remote Sens. Environ. 2009, 113, 2345–2355. [Google Scholar] [CrossRef]
- Franklin, J. Mapping Species Distributions: Spatial Inference and Prediction; Usher, M., Saunders, D., Peet, R., Dobson, A., Eds.; Cambridge University Press: Cambridge, UK, 2009; ISBN 9780521700023. [Google Scholar]
- Holmgren, J.; Persson, Å. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Naesset, E. Airborne laser scanning as a method in operational forest inventory: Status of accuracy assessments accomplished in Scandinavia. Scand. J. For. Res. 2007, 22, 433–442. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Korpela, I.; Maltamo, M.; Tokola, T. Imputation of single-tree attributes using airborne laser scanning-based height, intensity, and alpha shape metrics. Remote Sens. Environ. 2010, 114, 1263–1276. [Google Scholar] [CrossRef]
- Kim, S.; Hinckley, T.; Briggs, D. Classifying individual tree genera using stepwise cluster analysis based on height and intensity metrics derived from airborne laser scanner data. Remote Sens. Environ. 2011, 115, 3329–3342. [Google Scholar] [CrossRef]
- Kavzoglu, T.; Mather, P.M. The role of feature selection in artificial neural network applications. Int. J. Remote Sens. 2002, 23, 2919–2937. [Google Scholar] [CrossRef]
- Murtaugh, P.A. Performance of several variable-selection methods applied to real ecological data. Ecol. Lett. 2009, 12, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Kulasekera, K.B. Variable selection by stepwise slicing in nonparametric regression. Stat. Probab. Lett. 2001, 51, 327–336. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regression and variable selection via the elastic net. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Popescu, S.; Mallick, B.; Valle, D.; Zhao, K.; Zhang, X. Hyperspectral remote sensing of plant biochemistry using Bayesian model averaging with variable and band selection. Remote Sens. Environ. 2013, 132, 102–119. [Google Scholar]
- Packalén, P.; Temesgen, H.; Maltamo, M. Variable selection strategies for nearest neighbor imputation methods used in remote sensing based forest inventory. Can. J. Remote Sens. 2012, 38, 557–569. [Google Scholar] [CrossRef]
- Rakotomamonjy, A. Variable Selection Using SVM-based Criteria. J. Mach. Learn. Res. 2003, 3, 1357–1370. [Google Scholar]
- Genuer, R.; Poggi, J.M.; Tuleau-Malot, C. Variable selection using random forests. Pattern Recognit. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef] [Green Version]
- Demsar, J.; Curk, T.; Erjavec, A.; Gorup, C.; Hocevar, T.; Milutinovic, M.; Mozina, M.; Polajnar, M.; Toplak, M.; Staric, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Montesano, P.M.; Cook, B.D.; Sun, G.; Simard, M.; Nelson, R.F.; Ranson, K.J.; Zhang, Z.; Luthcke, S. Achieving accuracy requirements for forest biomass mapping: A spaceborne data fusion method for estimating forest biomass and LiDAR sampling error. Remote Sens. Environ. 2013, 130, 153–170. [Google Scholar] [CrossRef]
- Evans, J.S.; Hudak, A.T.; Faux, R.; Smith, A.M.S. Discrete return lidar in natural resources: Recommendations for project planning, data processing, and deliverables. Remote Sens. 2009, 1, 776–794. [Google Scholar] [CrossRef]
- Duncanson, L.I.; Dubayah, R.O.; Cook, B.D.; Rosette, J.; Parker, G. The importance of spatial detail: Assessing the utility of individual crown information and scaling approaches for lidar-based biomass density estimation. Remote Sens. Environ. 2015, 168, 102–112. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Remote estimation of chlorophyll content in higher plant leaves. Int. J. Remote Sens. 1997, 18, 2691–2697. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Zur, Y.; Chivkunova, O.B.; Merzlyak, M.N. Assessing Carotenoid Content in Plant Leaves with Reflectance Spectroscopy. Photochem. Photobiol. 2002, 75, 272–281. [Google Scholar] [CrossRef]
- Datt, B. Visible/near infrared reflectance and chlorophyll content in eucalyptus leaves. Int. J. Remote Sens. 1999, 20, 2741–2759. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation; NASA: Greenbelt, MD, USA, 1979.
- Zarco-Tejada, P.J.; Berjón, A.; López-Lozano, R.; Miller, J.R.; Martín, P.; Cachorro, V.; González, M.R.; De Frutos, A. Assessing vineyard condition with hyperspectral indices: Leaf and canopy reflectance simulation in a row-structured discontinuous canopy. Remote Sens. Environ. 2005, 99, 271–287. [Google Scholar] [CrossRef]
- Maccioni, A.; Agati, G.; Mazzinghi, P. New vegetation indices for remote measurement of chlorophylls based on leaf directional reflectance spectra. J. Photochem. Photobiol. B Biol. 2001, 61, 52–61. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of Vegetation Indices and a Modified Simple Ratio for Boreal Applications. Can. J. Remote Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures, and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W.; Harlan, J.C. Monitoring the Vernal Advancements and Retrogradation of Natural Vegetation; Texas A & M University: College Station, TX, USA, 1974. [Google Scholar]
- Barnes, J.D.; Balaguer, L.; Manrique, E.; Elvira, S.; Davison, A.W. A Reappraisal of the use of DMSO for the extraction and determination of chlorophylls a and b in lichens and other plants. Environ. Exp. Bot. 1992, 32, 85–100. [Google Scholar] [CrossRef]
- Gamon, J.A.; Peñuelas, J.; Field, C.B. A Narrow-Waveband Spectral Index That Tracks Diurnal Changes in Photosynthetic Efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Roujean, J.; Breon, F. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2000, 76, 156–172. [Google Scholar] [CrossRef]
- Gamon, J.A.; Surfus, J.S. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 1999, 143, 105–117. [Google Scholar] [CrossRef] [Green Version]
- Penuelas, J.; Baret, F.; Filella, I. Semi-empirical indices to assess carotenoids/chlorophyll a ratio from leaf spectral reflectance. Photosynthetica 1995, 31, 221–230. [Google Scholar]
- Vogelmann, J.E.; Rock, B.N.; Moss, D.M. Red edge spectral measurements from sugar maple leaves. Int. J. Remote Sens. 1993, 14, 1563–1575. [Google Scholar] [CrossRef]
- Northern Research Station Howland Cooperating Experimental Forest. Available online: https://www.nrs.fs.fed.us/ef/locations/me/howland_cef/ (accessed on 12 March 2019).
- Seymour, R.; Crandall, M.; Rogers, N.; Kenefic, L.; Sendak, P. Sixty Years of Silviculture in a Northern Conifer Forest in Maine, USA. For. Sci. 2017, 64, 102–111. [Google Scholar]
- Brissette, J.C.; Kenefic, L.S. History of the Penobscot Experimental Forest, 1950–2010; Forest Service, Northern Research Station: Newtown Square, PA, USA, 2014. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Hernandez-Orallo, J.; Flach, P.; Ferri, C. Brier Curves: A New Cost-Based Visualisation of Classifier Performance. In Proceedings of the the 28th International Conference on Machine Learning (ICML-11), Bellevue, WA, USA, 28 June–2 July 2011; pp. 1–27. [Google Scholar]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Congalton, R.G.; Green, K. Assessing the Accuracy of Remotely Sensed Data: Principles and Practices, 2nd ed.; CRC Press, Taylor and Francis Group: Boca Raton, FL, USA, 2009. [Google Scholar]
- Anderson, J.E.; Plourde, L.C.; Martin, M.E.; Braswell, B.H.; Smith, M.L.; Dubayah, R.O.; Hofton, M.A.; Blair, J.B. Integrating waveform lidar with hyperspectral imagery for inventory of a northern temperate forest. Remote Sens. Environ. 2008, 112, 1856–1870. [Google Scholar] [CrossRef]
- Ducic, V.; Hollaus, M.; Ullrich, A.; Wagner, W.; Melzer, T. 3D Vegetation Mapping And Classification Using Full-Wavevorm Laser Scanning. In Proceedings of the International Workshop 3D Remote Sensing in Forestry, Vienna, Austria, 14–15 February 2006; Volume Session 8a, pp. 211–217. [Google Scholar]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Guo, Q.; Rundel, P.W. Measuring dominance and diversity in ecological communities: Choosing the right variables. J. Veg. Sci. 1997, 8, 405–408. [Google Scholar] [CrossRef]
- Korpela, I.; Orka, H.O.; Maltamo, M.; Tokola, T.; Hyyppa, J. Tree Species Classification Using Airborne LiDAR-Effects of Stand and Tree Parameters, Downsizing of Training Set, Intensity Normalization, and Sensor Type. Silva Fenn. 2010, 44, 319–339. [Google Scholar] [CrossRef]
- Drake, J.B.; Dubayah, R.O.; Clark, D.B.; Knox, R.G.; Blair, J.B.; Hofton, M.A.; Chazdon, R.L.; Weishampel, J.F.; Prince, S. Estimation of tropical forest structural characteristics, using large-footprint lidar. Remote Sens. Environ. 2002, 79, 305–319. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. Support vector machines for classification in remote sensing. Int. J. Remote Sens. 2005, 26, 1007–1011. [Google Scholar] [CrossRef]
- Benediktsson, J.A.; Swain, P.H.; Erosy, O.K. Methods in Classification of Multisource. IEEE Trans. Geosci. Remote Sens. 1990, 28, 540–552. [Google Scholar]
- Haykin, S. Neural Networks: A Comprehensive Foundation, 2nd ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2004. [Google Scholar]
- Clark, P.; Niblett, T. The CN2 induction algorithm. Mach. Learn. 1989, 3, 261. [Google Scholar] [CrossRef]
- Brennan, R.; Webster, T.L. Object-oriented land cover classification of lidar-derived surfaces. Can. J. Remote Sens. 2006, 32, 162–172. [Google Scholar] [CrossRef]





| Name and Description of Metric | Units | Abbreviation |
|---|---|---|
| Mean absolute deviation = mean (|height − mean height|) of tree returns | Meters | AAD |
| Canopy relief ratio = ((mean − min)/(max − min)) of tree returns | Unitless | CRR |
| Density deciles of tree returns (number of returns in 10% height bins/total LiDAR returns) | Fraction | D0–D9 |
| Fraction of first return pulses intercepted by tree | Fraction | FCover |
| Fraction of all returns classified as tree | Fraction | FractAll |
| Interquartile range (P75–P25) of tree returns | Meters | IQR |
| Kurtosis of tree return heights | Meters | Kurt |
| Median absolute deviation = median (|height − median height|) of tree returns | Meters | MAD |
| Mean of tree return heights | Meters | Mean |
| Height percentiles (10% increments) of tree returns | Meters | P10–P100 |
| Quadratic mean of tree return heights | Meters | QMean |
| Skewness of tree return heights | Meters | Skew |
| Standard deviation of tree return heights | Meters | StDev |
| Vertical distribution ratio = (P100 − P50)/P100 | Unitless | VDR |
| Name of Vegetation Index | Abbreviation | Reference |
|---|---|---|
| Anthocyanin reflectance index 1 | ARI1 | [65] |
| Anthocyanin reflectance index 2 | ARI2 | [65] |
| Carotenoid reflectance index 2 | CRI2 | [66] |
| Datt 1 | DATT1 | [67] |
| Datt 2 | DATT2 | [67] |
| Difference vegetation index | DVI | [68] |
| Greenness index | GI | [69] |
| Gitelson and Merzlyak 1 | GM1 | [65] |
| Gitelson and Merzlyak 2 | GM2 | [65] |
| Maccioni | MAC | [70] |
| Modified simple ratio | MSR | [71] |
| Medium Resolution Imaging Spectrometer (MERIS) terrestrial chlorophyll index | MTCI | [72] |
| Modified triangular vegetation index | MTVI | [73] |
| Modified triangular vegetation index 2 | MTVI2 | [73] |
| Modified red edge normalized difference vegetation index | MRENDVI | [74] |
| Normalized difference vegetation index | NDVI | [75] |
| Normalized phaeophytinization index | NPQI | [76] |
| Photochemical reflectance index | PRI | [77] |
| Renormalized difference vegetation index | RDVI | [78] |
| Red edge inflection point | REIP | [79] |
| Red green ratio index | RGRI | [80] |
| Structure insensitive pigment index | SIPI | [81] |
| Vogelmann | VOG | [82] |
| Abbreviation | Common and Latin Names | Forest Where Dominant |
|---|---|---|
| ABBA | Balsam fir (Abies balsamea) | H, P |
| ACRU | Red maple (Acer rubrum) | H, P |
| ACSA | Silver maple (Acer saccharinum) | P |
| ACSP | Mountain maple (Acer spicatum) | P |
| BEAL | Yellow birch (Betula alleghaniensis) | P |
| BEPA | Paper birch (Betula papyrifera) | P |
| BEPO | Gray birch (Betula populifolia) | P |
| FAGR | American beech (Fagus grandifolia) | H, P |
| FRAM | White ash (Fraxinus americana) | H |
| FRPE | Green ash (Fraxinus pennsylvanica) | |
| LALA | Tamarack (Larix laricina) | |
| OSVI | Eastern hophornbeam (Ostrya virginiana) | |
| PIAB | Norway spruce (Picea abies) | H |
| PIMA | Black spruce (Picea mariana) | H |
| PIRE | Red pine (Pinus resinosa) | P |
| PIRU | Red spruce (Picea rubens) | P |
| PIST | Eastern white pine (Pinus strobus) | H, P |
| POBA | Balsam poplar (Populus balsamifera) | |
| POGR | Bigtooth aspen (Populus grandidentata) | P |
| POTR | Quaking aspen (Populus tremuloides) | P |
| QURU | Northern red oak (Quercus rubra) | |
| TIAM | American basswood (Tilia americana) | |
| THOC | Northern white cedar (Thuja occidentalis) | H, P |
| TSCA | Eastern hemlock (Tsuga canadensis) | H, P |
| ULAM | American elm (Ulmus americana) |
| Penobscot Experimental Forest | Howland Experimental Forest | ||||||
|---|---|---|---|---|---|---|---|
| LiDAR Metrics | Refl. Bands | Veg. Indices | LiDAR + Refl. + VIs | LiDAR Metrics | Refl. Bands | Veg. Indices | LiDAR + Refl. + VIs |
| D0 | 426.8 nm | ARI1 | 558.8 nm | D3 | 426.8 nm | ARI1 | 514.8 nm |
| D2 | 484.0 nm | ARI2 | 563.2 nm | D5 | 497.2 nm | ARI2 | 563.2 nm |
| D9 | 545.6 nm | DATT1 | 576.4 nm | D6 | 528.0 nm | CRI2 | DATT2 |
| FCover | 572.0 nm | DVI | 594.0 nm | D9 | 576.4 nm | DATT2 | FractAll |
| FractAll | 580.8 nm | GM1 | ARI2 | FCover | 580.8 nm | GI | GM1 |
| Kurtosis | 602.8 nm | MAC | CRI2 | FractAll | 651.2 nm | GM2 | GM2 |
| Mean | 655.6 nm | MTCI | D4 | P50 | 673.2 nm | PRI | Mean |
| P10 | 660.0 nm | MTVI | D6 | P60 | 686.4 nm | RDVI | MTCI |
| P40 | 673.2 nm | MTVI2 | GM1 | P90 | 690.8 nm | RGRI | P60 |
| P50 | 800.8 nm | MRENDVI | MSR | P100 | 721.6 nm | SIPI | P70 |
| P80 | 888.8 nm | NDVI | MTCI | 730.4 nm | PRI | ||
| P100 | 906.4 nm | NPQI | NPQI | 761.2 nm | REIP | ||
| StDev | 915.2 nm | PRI | P20 | 792.0 nm | RGRI | ||
| REIP | P100 | 862.4 nm | SIPI | ||||
| SIPI | RDVI | 906.4 nm | VOG | ||||
| StDev | 910.8 nm | ||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marrs, J.; Ni-Meister, W. Machine Learning Techniques for Tree Species Classification Using Co-Registered LiDAR and Hyperspectral Data. Remote Sens. 2019, 11, 819. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11070819
Marrs J, Ni-Meister W. Machine Learning Techniques for Tree Species Classification Using Co-Registered LiDAR and Hyperspectral Data. Remote Sensing. 2019; 11(7):819. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11070819
Chicago/Turabian StyleMarrs, Julia, and Wenge Ni-Meister. 2019. "Machine Learning Techniques for Tree Species Classification Using Co-Registered LiDAR and Hyperspectral Data" Remote Sensing 11, no. 7: 819. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11070819