Preliminary Application of Ground-Penetrating Radar for Reconstruction of Root System Architecture in Moso Bamboo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Experimental Design
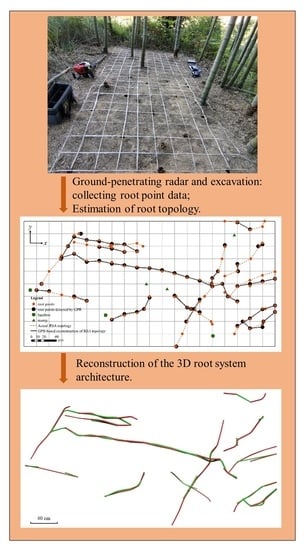
2.2. Radar Analysis
2.3. 3D RSA Reconstruction
- scoring (): Due to the insignificant change in diameter in mature rhizomes, () is divided into five classes, such that higher values of are represented by lower classes. A score of ten represents the highest class, with subsequent classes represented by subtracting increments of two from the score. If , the score is zero.
- scoring (): Similarly, () is divided into ten classes, with higher classes representing lower values of . A score of ten represents the highest class, with one subtracted from the score for each class below this maximum score. If , the score is zero.
- scoring (): Due to the insignificant variation in rhizome growth direction, changes in rhizome growth direction between 0° and 45° are divided into ten classes, such that every increment of 4.5° represents a score class. Higher values of are represented by lower classes. A score of ten represents the highest class, with one subtracted from the score for each subsequent class. If , the score assigned is −4, which counteracts the contribution of the and scores to the overall root point score function.
- Determine the starting root point. Arrange all root points in ascending order based on x-direction. The first occurring root point serves as the initial root point of the root system;
- Establish candidate root points. Calculate the distance between each root point and the starting root point in the x-direction () and the y-direction (). Search along in the x-direction from the origin. If ≤ 20 cm and ≤ 20 cm, these points are included as candidate root points;
- Determine the (n ≥ 2) root point. The score of each candidate root point is calculated using the score function. The root point with the highest score is considered the best-connected root point, while the other root points are involved in the reconstruction of the next root system topology. If the highest score is held by more than one root point, the root point with the lowest is considered to be the best-connected. The score function () for each candidate point is as follows:where n is the root point, and a, b, and c are the weight parameters. Note that is not used to calculate the score of the second candidate root point;
- Repeat step three until the reconstruction of the topology of the root system is complete. It should be noted that the root point identified as the best-connected of this root system is no longer involved in the reconstruction of the next root system;
- Repeat steps one to four until all root system topologies are reconstructed.
2.4. Evaluation of the 3D RSA Reconstructed
3. Results
3.1. GPR Detection of Rhizomes
3.2. RSA Reconstructed
4. Discussion
4.1. Application of GPR to Rhizome Systems
4.2. Limitations of RSA Reconstruction Based on GPR
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Smet, I.; White, P.J.; Bengough, A.G.; Dupuy, L.; Parizot, B.; Casimiro, I.; Heidstra, R.; Laskowski, M.; Lepetit, M.; Hochholdinger, F.; et al. Analyzing lateral root development: How tomove forward. Plant Cell 2012, 24, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Nicoll, B.C.; Ray, D. Adaptive growth of tree root systems in response to wind action and site conditions. Tree Physiol. 1996, 16, 891–898. [Google Scholar] [CrossRef] [Green Version]
- Kell, D.B. Breeding crop plants with deep roots: Their role in sustainable carbon, nutrient and water sequestration. Ann. Bot. 2011, 108, 407–418. [Google Scholar] [CrossRef] [Green Version]
- Kell, D.B. Large-scale sequestration of atmospheric carbon via plant roots in natural and agricultural ecosystems: Why and how. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1589–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resh, S.C.; Battaglia, M.; Worledge, D.; Ladiges, S. Coarse root biomass for eucalypt plantations in Tasmania, Australia: Sources of variation and methods for assessment. Trees Struct. Funct. 2003, 17, 389–399. [Google Scholar] [CrossRef]
- Sorgonà, A.; Proto, A.R.; Abenavoli, L.M.; Di Iorio, A. Spatial distribution of coarse root biomass and carbon in a high-density olive orchard: Effects of mechanical harvesting methods. Trees Struct. Funct. 2018, 32, 919–931. [Google Scholar] [CrossRef]
- Takahashi, H.; Pradal, C. Root phenotyping: Important and minimum information required for root modeling in crop plants. Breed. Sci. 2021, 71, 109–116. [Google Scholar] [CrossRef]
- Piñeros, M.A.; Larson, B.G.; Shaff, J.E.; Schneider, D.J.; Falcão, A.X.; Yuan, L.; Clark, R.T.; Craft, E.J.; Davis, T.W.; Pradier, P.-L.; et al. Evolving technologies for growing, imaging and analyzing 3D root system architecture of crop plants. J. Integr. Plant Biol. 2016, 58, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Langridge, P. Breeding Technologies to Increase Crop Production in a Changing World. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.R.; Shen, J.B.; Liao, H. Acquisition or utilization, which is more critical for enhancing phosphorus efficiency in modern crops? Plant Sci. 2010, 179, 302–306. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, J.; Zheng, B.Y.; Guo, Y. Optimizing soil-coring strategies to quantify root-length-density distribution in field-grown maize: Virtual coring trials using 3D root architecture models. Ann. Bot. 2018, 121, 809–819. [Google Scholar] [CrossRef]
- Zhu, J.M.; Ingram, P.A.; Benfey, P.N.; Elich, T. From lab to field, new approaches to phenotyping root system architecture. Curr. Opin. Plant Biol. 2011, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Buczko, U.; Kuchenbuch, R.O.; Gerke, H.H. Evaluation of a core sampling scheme to characterize root length density of maize. Plant Soil 2008, 316, 205. [Google Scholar] [CrossRef]
- Cahill, J.F.; McNickle, G.G.; Haag, J.J.; Lamb, E.G.; Nyanumba, S.M.; Clair, C.C.S. Plants Integrate Information about Nutrients and Neighbors. Science 2010, 328, 1657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reubens, B.; Poesen, J.; Danjon, F.; Geudens, G.; Muys, B. The role of fine and coarse roots in shallow slope stability and soil erosion control with a focus on root system architecture: A review. Trees Struct. Funct. 2007, 21, 385–402. [Google Scholar] [CrossRef]
- Danjon, F.; Fourcaud, T.; Bert, D. Root architecture and wind-firmness of mature Pinus pinaster. New Phytol. 2005, 168, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Godin, C.; Costes, E.; Sinoquet, H. A Method for Describing Plant Architecture Which Integrates Topology and Geometry. Ann. Bot. 1999, 84, 343–357. [Google Scholar] [CrossRef]
- Gärtner, H.; Wagner, B.; Heinrich, I.; Denier, C. 3D-laser scanning: A new method to analyze coarse tree root systems. For. Snow Landsc. Res. 2009, 82, 95–106. [Google Scholar]
- Zenone, T.; Morelli, G.; Teobaldelli, M.; Fischanger, F.; Matteucci, M.; Sordini, M.; Armani, A.; Ferrè, C.; Chiti, T.; Seufert, G. Preliminary use of ground-penetrating radar and electrical resistivity tomography to study tree roots in pine forests and poplar plantations. Funct. Plant Biol. 2008, 35, 1047–1058. [Google Scholar] [CrossRef]
- Song, X.Z.; Zhou, G.M.; Jiang, H.; Yu, S.Q.; Fu, J.H.; Li, W.Z.; Wang, W.F.; Ma, Z.H.; Peng, C.H. Carbon sequestration by Chinese bamboo forests and their ecological benefits: Assessment of potential, problems, and future challenges. Environ. Rev. 2011, 19, 418–428. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Cai, Y.; Xiao, L.D.; Gao, X.Y.; Shi, Y.J.; Du, H.Q.; Zhou, Y.F.; Zhou, G.M. Effects of different planting approaches and site conditions on aboveground carbon storage along a 10-year chronosequence after moso bamboo reforestation. For. Ecol. Manag. 2021, 482, 118867. [Google Scholar] [CrossRef]
- Xu, L.; Fang, H.Y.; Deng, X.; Ying, J.Y.; Lv, W.J.; Shi, Y.J.; Zhou, G.M.; Zhou, Y.F. Biochar application increased ecosystem carbon sequestration capacity in a Moso bamboo forest. For. Ecol. Manag. 2020, 475, 118447. [Google Scholar] [CrossRef]
- Yen, T.M.; Lee, J.S. Comparing aboveground carbon sequestration between moso bamboo (Phyllostachys heterocycla) and China fir (Cunninghamia lanceolata) forests based on the allometric model. For. Ecol. Manag. 2011, 261, 995–1002. [Google Scholar] [CrossRef]
- Isagi, Y.; Oda, T.; Fukushima, K.; Lian, C.; Yokogawa, M.; Kaneko, S. Predominance of a single clone of the most widely distributed bamboo species Phyllostachys edulis in East Asia. J. Plant Res. 2016, 129, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.M.; Mao, S.Y.; Wang, L.F.; Ye, X.H.; Wu, J.; Wang, G.R.; Chen, F.S.; Yang, Q.P. Clonal integration driven by source-sink relationships is constrained by rhizome branching architecture in a running bamboo species (Phyllostachys glauca): A 15N assessment in the field. For. Ecol. Manag. 2021, 481, 118754. [Google Scholar] [CrossRef]
- Li, Y.; Chen, J.; Xue, G.; Peng, Y.; Song, H. Effect of clonal integration on nitrogen cycling in rhizosphere of rhizomatous clonal plant, Phyllostachys bissetii, under heterogeneous light. Sci. Total Environ. 2018, 628–629, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Chen, J.; Cui, X.H.; Fan, B.H.; Lin, H. Application of ground penetrating radar for coarse root detection and quantification: A review. Plant Soil 2013, 362. [Google Scholar] [CrossRef] [Green Version]
- Lontoc-Roy, M.; Dutilleul, P.; Prasher, S.O.; Han, L.; Smith, D.L. Computed tomography scanning for three-dimensional imaging and complexity analysis of developing root systems. Can. J. Bot. 2005, 83, 1434–1442. [Google Scholar] [CrossRef]
- Barton, C.V.M.; Montagu, K.D. Detection of tree roots and determination of root diameters by ground penetrating radar under optimal conditions. Tree Physiol. 2004, 24, 1323–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butnor, J.R.; Doolittle, J.A.; Johnsen, K.H.; Samuelson, L.; Stokes, T.; Kress, L. Utility of ground-penetrating radar as a root biomass survey tool in forest systems. Soil Sci. Soc. Am. J. 2003, 67, 1607–1615. [Google Scholar] [CrossRef] [Green Version]
- Butnor, J.R.; Doolittle, J.A.; Kress, L.; Cohen, S.; Johnsen, K.H. Use of ground-penetrating radar to study tree roots in the Southeastern United States. Tree Physiol. 2001, 21, 1269–1278. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.H.; Chen, J.; Shen, J.S.; Cao, X.; Chen, X.H.; Zhu, X.L. Modeling tree root diameter and biomass by ground-penetrating radar. Sci. China Earth Sci. 2011, 54, 711–719. (In Chinese) [Google Scholar] [CrossRef]
- Cui, X.H.; Zhang, Z.; Guo, L.; Liu, X.B.; Quan, Z.X.; Cao, X.; Chen, X.H. The Root-Soil Water Relationship Is Spatially Anisotropic in Shrub-Encroached Grassland in North China: Evidence from GPR Investigation. Remote Sens. 2021, 13, 1137. [Google Scholar] [CrossRef]
- Guo, L.; Wu, Y.; Chen, J.; Hirano, Y.; Tanikawa, T.; Li, W.T.; Cui, X.H. Calibrating the impact of root orientation on root quantification using ground-penetrating radar. Plant Soil 2015, 395, 289–305. [Google Scholar] [CrossRef]
- Hruska, J.; Čermák, J.; Šustek, S. Mapping tree root systems with ground-penetrating radar. Tree Physiol. 1999, 19, 125–130. [Google Scholar] [CrossRef] [Green Version]
- Isaac, M.E.; Anglaaere, L.C.N. An in situ approach to detect tree root ecology: Linking ground-penetrating radar imaging to isotope-derived water acquisition zones. Ecol. Evol. 2013, 3, 1330–1339. [Google Scholar] [CrossRef]
- Stokes, A.; Fourcaud, T.; Hruska, J.; Cermak, J.; Nadyezdhina, N.; Praus, L. An evaluation of different methods to investigate root system architecture of urban trees in situ: I. ground-penetrating radar. J. Arboric. 2002, 28, 2–10. [Google Scholar]
- Wu, Y.; Guo, L.; Cui, X.; Chen, J.; Cao, X.; Lin, H. Ground-penetrating radar-based automatic reconstruction of three-dimensional coarse root system architecture. Plant Soil 2014, 383, 155–172. [Google Scholar] [CrossRef]
- Ohashi, M.; Ikeno, H.; Sekihara, K.; Tanikawa, T.; Dannoura, M.; Yamase, K.; Todo, C.; Tomita, T.; Hirano, Y. Reconstruction of root systems in Cryptomeria japonica using root point coordinates and diameters. Planta 2019, 249, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Hirano, Y.; Dannoura, M.; Aono, K.; Igarashi, T.; Ishii, M.; Yamase, K.; Makita, N.; Kanazawa, Y. Limiting factors in the detection of tree roots using ground-penetrating radar. Plant Soil 2009, 319, 15–24. [Google Scholar] [CrossRef]
- Liu, X.B.; Cui, X.H.; Guo, L.; Chen, J.; Li, W.T.; Yang, D.; Di Cao, X.; Chen, X.H.; Liu, Q.X.; Lin, H. Non-invasive estimation of root zone soil moisture from coarse root reflections in ground-penetrating radar images. Plant Soil 2019, 436, 623–639. [Google Scholar] [CrossRef]
- Ristic, A.V.; Petrovacki, D.; Govedarica, M. A new method to simultaneously estimate the radius of a cylindrical object and the wave propagation velocity from GPR data. Comput. Geosci. 2009, 35, 1620–1630. [Google Scholar] [CrossRef]
- Fan, H.H. Study on Rhizome Growth Regularity of New-planted Phyllostachys edulis. J. Fujian Coll. For. 1999, 19, 30–32. (In Chinese) [Google Scholar]
- Jiang, G.H.; Yu, L.H.; Li, Z.D.; Niu, H.W.; Shi, L. Hierarchical system and its quantitative attribute of moso bamboo rhizome. Chin. J. Ecol. 2017, 36, 3479–3484. (In Chinese) [Google Scholar] [CrossRef]
- Zhu, S.P.; Huang, C.L.; Su, Y.; Sato, M. 3D Ground Penetrating Radar to Detect Tree Roots and Estimate Root Biomass in the Field. Remote Sens. 2014, 6, 5754–5773. [Google Scholar] [CrossRef] [Green Version]
- Hirano, Y.; Yamamoto, R.; Dannoura, M.; Aono, K.; Igarashi, T.; Ishii, M.; Yamase, K.; Makita, N.; Kanazawa, Y. Detection frequency of Pinus thunbergii roots by ground-penetrating radar is related to root biomass. Plant Soil 2012, 360, 363–373. [Google Scholar] [CrossRef]
- Tanikawa, T.; Ikeno, H.; Dannoura, M.; Yamase, K.; Aono, K.; Hirano, Y. Leaf litter thickness, but not plant species, can affect root detection by ground penetrating radar. Plant Soil 2016, 408, 271–283. [Google Scholar] [CrossRef]
- Chen, X.X.; He, R.Y.; Ding, Q.S.; Sun, Q. A Digitization and Visualization Procedure for 3D Wheat Root System Architecture in Rice-Wheat Rotation. J. Inst. Eng. Ser. A 2019, 100, 1–8. [Google Scholar] [CrossRef]
- Danjon, F.; Reubens, B. Assessing and analyzing 3D architecture of woody root systems, a review of methods and applications in tree and soil stability, resource acquisition and allocation. Plant Soil 2008, 303, 1–34. [Google Scholar] [CrossRef]
- Liu, Q.X.; Cui, X.H.; Liu, X.B.; Chen, J.; Chen, X.H.; Cao, X. Detection of Root Orientation Using Ground-Penetrating Radar. IEEE Trans. Geosci. Remote Sens. 2018, 56, 93–104. [Google Scholar] [CrossRef]
- Tanikawa, T.; Hirano, Y.; Dannoura, M.; Yamase, K.; Aono, K.; Ishii, M.; Igarashi, T.; Ikeno, H.; Kanazawa, Y. Root orientation can affect detection accuracy of ground-penetrating radar. Plant Soil 2013, 373, 317–327. [Google Scholar] [CrossRef]
- Bain, J.; Day, F.; Butnor, J. Experimental Evaluation of Several Key Factors Affecting Root Biomass Estimation by 1500 MHz Ground-Penetrating Radar. Remote Sens. 2017, 9, 1337. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Lin, H.; Fan, B.H.; Cui, X.H.; Chen, J. Impact of root water content on root biomass estimation using ground penetrating radar: Evidence from forward simulations and field controlled experiments. Plant Soil 2013, 371, 503–520. [Google Scholar] [CrossRef]
- Dannoura, M.; Hirano, Y.; Igarashi, T.; Ishii, M.; Aono, K.; Yamase, K.; Kanazawa, Y. Detection of Cryptomeria japonica roots with ground penetrating radar. Plant Biosyst. 2008, 142, 375–380. [Google Scholar] [CrossRef]
- Yamase, K.; Tanikawa, T.; Dannoura, M.; Ohashi, M.; Todo, C.; Ikeno, H.; Aono, K.; Hirano, Y. Ground-penetrating radar estimates of tree root diameter and distribution under field conditions. Trees 2018, 32, 1657–1668. [Google Scholar] [CrossRef]
- De Coster, A.; Lambot, S. Fusion of Multifrequency GPR Data Freed from Antenna Effects. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 664–674. [Google Scholar] [CrossRef]
- Cui, X.; Liu, X.; Cao, X.; Fan, B.; Zhang, Z.; Chen, J.; Chen, X.; Lin, H.; Guo, L. Pairing dual-frequency GPR in summer and winter enhances the detection and mapping of coarse roots in the semi-arid shrubland in China. Eur. J. Soil Sci. 2020, 71, 236–251. [Google Scholar] [CrossRef]














| a | b | c | Accuracy (%) |
|---|---|---|---|
| 0.0 | 0.5 | 0.5 | 42.88 |
| 0.2 | 0.4 | 0.4 | 48.96 |
| 0.4 | 0.3 | 0.3 | 67.71 |
| 0.6 | 0.2 | 0.2 | 78.13 |
| 0.8 | 0.1 | 0.1 | 70.83 |
| 1.0 | 0.0 | 0.0 | 59.38 |
| Soil Depth (cm) | Soil Stone Content (%) | Soil Water Content (%) |
|---|---|---|
| 0–10 | 5.14 ± 2.29 | 10.84 ± 1.33 |
| 10–30 | 5.03 ± 1.89 | 10.29 ± 1.15 |
| 30–50 | 4.77 ± 2.34 | 9.30 ± 0.46 |
| Soil Depth (cm) | Measured Diameter (mm) | GPR-Based Estimated Diameter (mm) |
|---|---|---|
| 0–5 | 17.8 ± 3.6 a | 18.8 ± 4.3 a |
| 5–10 | 15.8 ± 3.4 a | 16.8 ± 3.7 a |
| 10–15 | 17.9 ± 2.3 a | 18.0 ± 2.2 a |
| 15–20 | 17.8 ± 4.3 a | 19.7 ± 2.6 a |
| ≥20 | 16.7 ± 0.6 a | 16.7 ± 2.1 a |
| Measured RSA | Reconstructed RSA | Accuracy (%) | GPR-Based Reconstructed | Accuracy (%) | |
|---|---|---|---|---|---|
| Total length (cm) | 1530.6 | 1457.9 | 91.98 | 986.5 | 64.45 |
| Total volume () | 3659.3 | 3483.9 | 95.21 | 2678.9 | 73.21 |
| Total biomass (g) | 2866.8 | 2715.7 | 94.73 | 2088.2 | 72.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, L.; Li, C.; Cai, Y.; Zhou, M.; Zhou, T.; Gao, X.; Du, H.; Zhou, Y.; Zhou, G. Preliminary Application of Ground-Penetrating Radar for Reconstruction of Root System Architecture in Moso Bamboo. Remote Sens. 2021, 13, 2816. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13142816
Xiao L, Li C, Cai Y, Zhou M, Zhou T, Gao X, Du H, Zhou Y, Zhou G. Preliminary Application of Ground-Penetrating Radar for Reconstruction of Root System Architecture in Moso Bamboo. Remote Sensing. 2021; 13(14):2816. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13142816
Chicago/Turabian StyleXiao, Longdong, Chong Li, Yue Cai, Mingxing Zhou, Tao Zhou, Xueyan Gao, Huaqiang Du, Yufeng Zhou, and Guomo Zhou. 2021. "Preliminary Application of Ground-Penetrating Radar for Reconstruction of Root System Architecture in Moso Bamboo" Remote Sensing 13, no. 14: 2816. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13142816