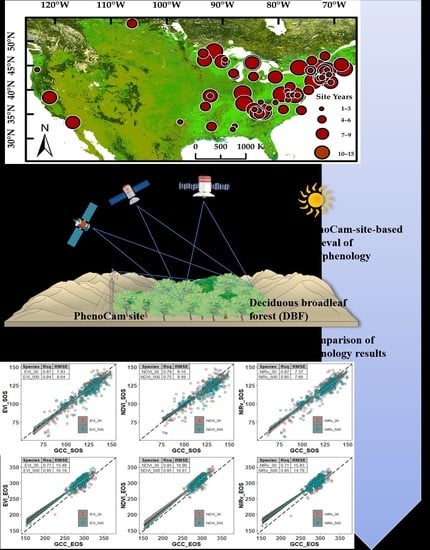
Comparing Different Spatial Resolutions and Indices for Retrieving Land Surface Phenology for Deciduous Broadleaf Forests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. PhenoCam Data
2.3. Satellite Remote Sensing Data
2.3.1. Landsat and Sentinel-2 Data
2.3.2. MODIS Data
2.4. Phenological Retrieval for Different Data
2.5. Comparisons of Phenological Results
3. Results
3.1. Time Series Variation of Satellite Vegetation Indices
3.2. Phenological Comparisons at Different Spatial Resolutions
3.3. Differences in GCC and Satellite Vegetation Indices for Different Phenological Metrics
4. Discussion
4.1. Applicability of Different Resolution Data (30 m and 500 m) for SOS and EOS
4.2. Gradient Buffers: Better Phenological Results?
4.3. Difficulties of Satellite Remote Sensing Monitoring of Phenology
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piao, S.; Wang, X.; Park, T.; Chen, C.; Lian, X.; He, Y.; Bjerke, J.W.; Chen, A.; Ciais, P.; Tømmervik, H.; et al. Characteristics, drivers and feedbacks of global greening. Nat. Rev. Earth Environ. 2020, 1, 14–27. [Google Scholar] [CrossRef]
- Hughes, L. Biological consequences of global warming: Is the signal already apparent? Trends Ecol. Evol. 2000, 15, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Liu, S.; Ma, K.; Zhu, Y. Relationships between soil characteristics, topography and plant diversity in a heterogeneous deciduous broad-leaved forest near Beijing, China. Plant Soil 2004, 261, 47–54. [Google Scholar] [CrossRef]
- Su, L.; Zhao, C.; Xu, W.; Xie, Z. Hydrochemical Fluxes in Bulk Precipitation, Throughfall, and Stemflow in a Mixed Evergreen and Deciduous Broadleaved Forest. Forests 2019, 10, 507. [Google Scholar] [CrossRef]
- Caparros-Santiago, J.A.; Rodriguez-Galiano, V.; Dash, J. Land surface phenology as indicator of global terrestrial ecosystem dynamics: A systematic review. ISPRS J. Photogramm. Remote Sens. 2021, 171, 330–347. [Google Scholar] [CrossRef]
- Zhang, X.; Friedl, M.A.; Schaaf, C.B.; Strahler, A.H.; Schneider, A. The footprint of urban climates on vegetation phenology. Geophys. Res. Lett. 2004, 31, L12209. [Google Scholar] [CrossRef]
- Cleland, E.E.; Chuine, I.; Menzel, A.; Mooney, H.A.; Schwartz, M.D. Shifting plant phenology in response to global change. Trends Ecol. Evol. 2007, 22, 357–365. [Google Scholar] [CrossRef]
- Heiderman, R.R.; Nettles, W.R.; Ontl, T.A.; Latimer, J.M.; Richardson, A.D.; Hanson, P.J. SPRUCE Manual Phenology Observations and Photographs Beginning in 2010; Oak Ridge National Lab. (ORNL): Oak Ridge, TN, USA, 2018.
- Deng, L.; Lin, Y.; Yan, L.; Tesfamichael, S.; Billen, R.; Yao, Y.; Yao, W.; Chen, X.; Fang, X.; Wang, C.; et al. Urban plant phenology monitoring: Expanding the functions of widespread surveillance cameras to nature rhythm understanding. Remote Sens. Appl. Soc. Environ. 2019, 15, 100232. [Google Scholar] [CrossRef]
- Zhang, C.; Xie, Z.A.; Shang, J.; Liu, J.; Dong, T.; Tang, M.; Feng, S.; Cai, H. Detecting winter canola (Brassica napus) phenological stages using an improved shape-model method based on time-series UAV spectral data. Crop. J. 2022, 10, 1353–1362. [Google Scholar] [CrossRef]
- Brown, T.B.; Hultine, K.R.; Steltzer, H.; Denny, E.G.; Denslow, M.W.; Granados, J.; Henderson, S.; Moore, D.; Nagai, S.; SanClements, M.; et al. Using phenocams to monitor our changing Earth: Toward a global phenocam network. Front. Ecol. Environ. 2016, 14, 84–93. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, X. Mapping Crop Phenology in Near Real-Time Using Satellite Remote Sensing: Challenges and Opportunities. J. Remote Sens. 2021, 2021, 8379391. [Google Scholar] [CrossRef]
- Berra, E.F.; Gaulton, R. Remote sensing of temperate and boreal forest phenology: A review of progress, challenges and opportunities in the intercomparison of in-situ and satellite phenological metrics. For. Ecol. Manag. 2021, 480, 118663. [Google Scholar] [CrossRef]
- Zhang, X.; Friedl, M.A.; Schaaf, C.B.; Strahler, A.H.; Hodges, J.C.F.; Gao, F.; Reed, B.C.; Huete, A. Monitoring vegetation phenology using MODIS. Remote Sens. Environ. 2003, 84, 471–475. [Google Scholar] [CrossRef]
- Cui, Y.; Xiao, X.; Dong, J.; Zhang, Y.; Qin, Y.; Doughty, R.B.; Wu, X.; Liu, X.; Joiner, J.; Moore, B., III. Continued Increases of Gross Primary Production in Urban Areas during 2000–2016. J. Remote Sens. 2022, 14, 2022. [Google Scholar] [CrossRef]
- Li, X.; Du, H.; Zhou, G.; Mao, F.; Zhang, M.; Han, N.; Fan, W.; Liu, H.; Huang, Z.; He, S.; et al. Phenology estimation of subtropical bamboo forests based on assimilated MODIS LAI time series data. ISPRS J. Photogramm. Remote Sens. 2021, 173, 262–277. [Google Scholar] [CrossRef]
- Touhami, I.; Moutahir, H.; Assoul, D.; Bergaoui, K.; Aouinti, H.; Bellot, J.; Andreu, J.M. Multi-year monitoring land surface phenology in relation to climatic variables using MODIS-NDVI time-series in Mediterranean forest, Northeast Tunisia. Acta Oecologica 2021, 114, 103804. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, S.; Feng, L.; Zhang, J.; Deng, F. Mapping Maize Cultivated Area Combining MODIS EVI Time Series and the Spatial Variations of Phenology over Huanghuaihai Plain. Appl. Sci. 2020, 10, 2667. [Google Scholar] [CrossRef]
- Leng, S.; Huete, A.; Cleverly, J.; Yu, Q.; Zhang, R.; Wang, Q. Spatiotemporal Variations of Dryland Vegetation Phenology Revealed by Satellite-Observed Fluorescence and Greenness across the North Australian Tropical Transect. Remote Sens. 2022, 14, 2985. [Google Scholar] [CrossRef]
- Jeong, S.-J.; Ho, C.-H.; Gim, H.-J.; Brown, M.E. Phenology shifts at start vs. end of growing season in temperate vegetation over the Northern Hemisphere for the period 1982–2008. Glob. Chang. Biol. 2011, 17, 2385–2399. [Google Scholar] [CrossRef]
- Jeong, S.-J.; Schimel, D.; Frankenberg, C.; Drewry, D.T.; Fisher, J.B.; Verma, M.; Berry, J.A.; Lee, J.-E.; Joiner, J. Application of satellite solar-induced chlorophyll fluorescence to understanding large-scale variations in vegetation phenology and function over northern high latitude forests. Remote Sens. Environ. 2017, 190, 178–187. [Google Scholar] [CrossRef]
- Walther, S.; Voigt, M.; Thum, T.; Gonsamo, A.; Zhang, Y.; Köhler, P.; Jung, M.; Varlagin, A.; Guanter, L. Satellite chlorophyll fluorescence measurements reveal large-scale decoupling of photosynthesis and greenness dynamics in boreal evergreen forests. Glob. Chang. Biol. 2016, 22, 2979–2996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, Y.; Dong, J.; Xiao, X. Green-up dates in the Tibetan Plateau have continuously advanced from 1982 to 2011. Proc. Natl. Acad. Sci. USA 2013, 110, 4309–4314. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Zhu, X.; Wan, L.; Collin, M. Impacts of Satellite Revisit Frequency on Spring Phenology Monitoring of Deciduous Broad-Leaved Forests Based on Vegetation Index Time Series. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 10500–10508. [Google Scholar] [CrossRef]
- Yang, J.; Xiao, X.; Doughty, R.; Zhao, M.; Zhang, Y.; Köhler, P.; Wu, X.; Frankenberg, C.; Dong, J. TROPOMI SIF reveals large uncertainty in estimating the end of plant growing season from vegetation indices data in the Tibetan Plateau. Remote Sens. Environ. 2022, 280, 113209. [Google Scholar] [CrossRef]
- Peng, D.; Wu, C.; Li, C.; Zhang, X.; Liu, Z.; Ye, H.; Luo, S.; Liu, X.; Hu, Y.; Fang, B. Spring green-up phenology products derived from MODIS NDVI and EVI: Intercomparison, interpretation and validation using National Phenology Network and AmeriFlux observations. Ecol. Indic. 2017, 77, 323–336. [Google Scholar] [CrossRef]
- Wagle, P.; Kakani, V.G.; Gowda, P.H.; Xiao, X.; Northup, B.K.; Neel, J.P.S.; Starks, P.J.; Steiner, J.L.; Gunter, S.A. Dormant Season Vegetation Phenology and Eddy Fluxes in Native Tallgrass Prairies of the U.S. Southern Plains. Remote Sens. 2022, 14, 2620. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, J.; Tong, X.; Zhang, J.; Meng, P.; Li, J.; Liu, P.; Yu, P. NIRv and SIF better estimate phenology than NDVI and EVI: Effects of spring and autumn phenology on ecosystem production of planted forests. Agric. For. Meteorol. 2022, 315, 108819. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, N.; Li, Y.; Li, Y. Sub-continental-scale mapping of tidal wetland composition for East Asia: A novel algorithm integrating satellite tide-level and phenological features. Remote Sens. Environ. 2022, 269, 112799. [Google Scholar] [CrossRef]
- Melaas, E.K.; Friedl, M.A.; Zhu, Z. Detecting interannual variation in deciduous broadleaf forest phenology using Landsat TM/ETM+ data. Remote Sens. Environ. 2013, 132, 176–185. [Google Scholar] [CrossRef]
- Ju, J.; Roy, D.P. The availability of cloud-free Landsat ETM+ data over the conterminous United States and globally. Remote Sens. Environ. 2008, 112, 1196–1211. [Google Scholar] [CrossRef]
- Junttila, S.; Ardö, J.; Cai, Z.; Jin, H.; Kljun, N.; Klemedtsson, L.; Krasnova, A.; Lange, H.; Lindroth, A.; Mölder, M.; et al. Estimating local-scale forest GPP in Northern Europe using Sentinel-2: Model comparisons with LUE, APAR, the plant phenology index, and a light response function. Sci. Remote Sens. 2023, 7, 100075. [Google Scholar] [CrossRef]
- Tian, J.; Zhu, X.; Wu, J.; Shen, M.; Chen, J. Coarse-Resolution Satellite Images Overestimate Urbanization Effects on Vegetation Spring Phenology. Remote Sens. 2020, 12, 117. [Google Scholar] [CrossRef]
- Bolton, D.K.; Gray, J.M.; Melaas, E.K.; Moon, M.; Eklundh, L.; Friedl, M.A. Continental-scale land surface phenology from harmonized Landsat 8 and Sentinel-2 imagery. Remote Sens. Environ. 2020, 240, 111685. [Google Scholar] [CrossRef]
- Moon, M.; Richardson, A.D.; Friedl, M.A. Multiscale assessment of land surface phenology from harmonized Landsat 8 and Sentinel-2, PlanetScope, and PhenoCam imagery. Remote Sens. Environ. 2021, 266, 112716. [Google Scholar] [CrossRef]
- Nagai, S.; Maeda, T.; Gamo, M.; Muraoka, H.; Suzuki, R.; Nasahara, K. Using digital camera images to detect canopy condition of deciduous broad-leaved trees. Plant Ecol. Divers. 2011, 4, 79–89. [Google Scholar] [CrossRef]
- Zhu, X.; Helmer, E.H.; Gwenzi, D.; Collin, M.; Fleming, S.; Tian, J.; Marcano-Vega, H.; Meléndez-Ackerman, E.J.; Zimmerman, J.K. Characterization of Dry-Season Phenology in Tropical Forests by Reconstructing Cloud-Free Landsat Time Series. Remote Sens. 2021, 13, 4736. [Google Scholar] [CrossRef]
- Thapa, S.; Garcia Millan, V.E.; Eklundh, L. Assessing Forest Phenology: A Multi-Scale Comparison of Near-Surface (UAV, Spectral Reflectance Sensor, PhenoCam) and Satellite (MODIS, Sentinel-2) Remote Sensing. Remote Sens. 2021, 13, 1597. [Google Scholar] [CrossRef]
- Richardson, A.D.; Hufkens, K.; Milliman, T.; Aubrecht, D.M.; Chen, M.; Gray, J.M.; Johnston, M.R.; Keenan, T.F.; Klosterman, S.T.; Kosmala, M.; et al. Tracking vegetation phenology across diverse North American biomes using PhenoCam imagery. Sci. Data 2018, 5, 180028. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jayavelu, S.; Liu, L.; Friedl, M.A.; Henebry, G.M.; Liu, Y.; Schaaf, C.B.; Richardson, A.D.; Gray, J. Evaluation of land surface phenology from VIIRS data using time series of PhenoCam imagery. Agric. For. Meteorol. 2018, 256, 137–149. [Google Scholar] [CrossRef]
- Seyednasrollah, B.; Young, A.M.; Hufkens, K.; Milliman, T.; Friedl, M.A.; Frolking, S.; Richardson, A.D. Tracking vegetation phenology across diverse biomes using Version 2.0 of the PhenoCam Dataset. Sci. Data 2019, 6, 222, Correction in Sci. Data 2019, 6, 261. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Deslauriers, A.; Morin, H.; Latifi, H.; Rossi, S. Comparing Time-Lapse PhenoCams with Satellite Observations across the Boreal Forest of Quebec, Canada. Remote Sens. 2022, 14, 100. [Google Scholar] [CrossRef]
- Zhao, W.; Qu, Y.; Zhang, L.; Li, K. Spatial-aware SAR-optical time-series deep integration for crop phenology tracking. Remote Sens. Environ. 2022, 276, 113046. [Google Scholar] [CrossRef]
- Moon, M.; Seyednasrollah, B.; Richardson, A.D.; Friedl, M.A. Using time series of MODIS land surface phenology to model temperature and photoperiod controls on spring greenup in North American deciduous forests. Remote Sens. Environ. 2021, 260, 112466. [Google Scholar] [CrossRef]
- Morin, X.; Lechowicz, M.J.; Augspurger, C.; O’Keefe, J.; Viner, D.; Chuine, I. Leaf phenology in 22 North American tree species during the 21st century. Glob. Chang. Biol. 2009, 15, 961–975. [Google Scholar] [CrossRef]
- Cui, T.; Martz, L.; Zhao, L.; Guo, X. Investigating the impact of the temporal resolution of MODIS data on measured phenology in the prairie grasslands. GISci. Remote Sens. 2020, 57, 395–410. [Google Scholar] [CrossRef]
- Liu, Y.; Bachofen, C.; Wittwer, R.; Duarte, G.S.; Sun, Q.; Klaus, V.H.; Buchmann, N. Using PhenoCams to track crop phenology and explain the effects of different cropping systems on yield. Agric. Syst. 2022, 195, 103306. [Google Scholar] [CrossRef]
- Filippa, G.; Cremonese, E.; Migliavacca, M.; Galvagno, M.; Sonnentag, O.; Humphreys, E.; Hufkens, K.; Ryu, Y.; Verfaillie, J.; di Cella, U.M.; et al. NDVI derived from near-infrared-enabled digital cameras: Applicability across different plant functional types. Agric. For. Meteorol. 2018, 249, 275–285. [Google Scholar] [CrossRef]
- Kowalski, K.; Senf, C.; Hostert, P.; Pflugmacher, D. Characterizing spring phenology of temperate broadleaf forests using Landsat and Sentinel-2 time series. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102172. [Google Scholar] [CrossRef]
- Zhang, H.K.; Roy, D.P.; Yan, L.; Li, Z.; Huang, H.; Vermote, E.; Skakun, S.; Roger, J.-C. Characterization of Sentinel-2A and Landsat-8 top of atmosphere, surface, and nadir BRDF adjusted reflectance and NDVI differences. Remote Sens. Environ. 2018, 215, 482–494. [Google Scholar] [CrossRef]
- Ahl, D.E.; Gower, S.T.; Burrows, S.N.; Shabanov, N.V.; Myneni, R.B.; Knyazikhin, Y. Monitoring spring canopy phenology of a deciduous broadleaf forest using MODIS. Remote Sens. Environ. 2006, 104, 88–95. [Google Scholar] [CrossRef]
- Yang, Y.; Luo, J.; Huang, Q.; Wu, W.; Sun, Y. Weighted Double-Logistic Function Fitting Method for Reconstructing the High-Quality Sentinel-2 NDVI Time Series Data Set. Remote Sens. 2019, 11, 2342. [Google Scholar] [CrossRef]
- Beck, P.S.; Atzberger, C.; Høgda, K.A.; Johansen, B.; Skidmore, A.K. Improved monitoring of vegetation dynamics at very high latitudes: A new method using MODIS NDVI. Remote Sens. Environ. 2006, 100, 321–334. [Google Scholar] [CrossRef]
- Atkinson, P.M.; Jeganathan, C.; Dash, J.; Atzberger, C. Inter-comparison of four models for smoothing satellite sensor time-series data to estimate vegetation phenology. Remote Sens. Environ. 2012, 123, 400–417. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, R.; Yin, G.; Wang, C.; Liu, G.; Verger, A.; Descals, A.; Filella, I.; Penuelas, J. Divergent Performances of Vegetation Indices in Extracting Photosynthetic Phenology for Northern Deciduous Broadleaf Forests. IEEE Geosci. Remote Sens. Lett. 2022, 19, 1–5. [Google Scholar] [CrossRef]
- Simonetti, D.; Simonetti, E.; Szantoi, Z.; Lupi, A.; Eva, H.D. First Results From the Phenology-Based Synthesis Classifier Using Landsat 8 Imagery. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1496–1500. [Google Scholar] [CrossRef]
- Vrieling, A.; Skidmore, A.K.; Wang, T.; Meroni, M.; Ens, B.J.; Oosterbeek, K.; O’Connor, B.; Darvishzadeh, R.; Heurich, M.; Shepherd, A.; et al. Spatially detailed retrievals of spring phenology from single-season high-resolution image time series. Int. J. Appl. Earth Obs. Geoinf. 2017, 59, 19–30. [Google Scholar] [CrossRef]
- Gray, R.E.J.; Ewers, R.M. Monitoring Forest Phenology in a Changing World. Forests 2021, 12, 297. [Google Scholar] [CrossRef]
- Peng, D.; Zhang, X.; Zhang, B.; Liu, L.; Liu, X.; Huete, A.R.; Huang, W.; Wang, S.; Luo, S.; Zhang, X.; et al. Scaling effects on spring phenology detections from MODIS data at multiple spatial resolutions over the contiguous United States. ISPRS J. Photogramm. Remote Sens. 2017, 132, 185–198. [Google Scholar] [CrossRef]
- Reed, B.C. Trend Analysis of Time-Series Phenology of North America Derived from Satellite Data. GISci. Remote Sens. 2006, 43, 24–38. [Google Scholar] [CrossRef]
- Zeng, H.; Jia, G.; Epstein, H. Recent changes in phenology over the northern high latitudes detected from multi-satellite data. Environ. Res. Lett. 2011, 6, 045508. [Google Scholar] [CrossRef]
- Yan, E.; Wang, G.; Lin, H.; Xia, C.; Sun, H. Phenology-based classification of vegetation cover types in Northeast China using MODIS NDVI and EVI time series. Int. J. Remote Sens. 2015, 36, 489–512. [Google Scholar] [CrossRef]
- Walker, J.J.; De Beurs, K.M.; Wynne, R.H.; Gao, F. Evaluation of Landsat and MODIS data fusion products for analysis of dryland forest phenology. Remote Sens. Environ. 2012, 117, 381–393. [Google Scholar] [CrossRef]
- Reid, C.E.; Kubzansky, L.D.; Li, J.; Shmool, J.L.; Clougherty, J.E. It’s not easy assessing greenness: A comparison of NDVI datasets and neighborhood types and their associations with self-rated health in New York City. Health Place 2018, 54, 92–101. [Google Scholar] [CrossRef]
- Su, J.G.; Dadvand, P.; Nieuwenhuijsen, M.J.; Bartoll, X.; Jerrett, M. Associations of green space metrics with health and behavior outcomes at different buffer sizes and remote sensing sensor resolutions. Environ. Int. 2019, 126, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Berra, E.F.; Gaulton, R.; Barr, S. Assessing spring phenology of a temperate woodland: A multiscale comparison of ground, unmanned aerial vehicle and Landsat satellite observations. Remote Sens. Environ. 2019, 223, 229–242. [Google Scholar] [CrossRef]
- Nakaji, T.; Oguma, H.; Hiura, T. Ground-based monitoring of the leaf phenology of deciduous broad-leaved trees using high resolution NDVI camera images. J. Agric. Meteorol. 2011, 67, 65–74. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, Y.; Tan, Z.; Song, Q.; Liang, N.; Yu, L.; Zhao, J. Using digital cameras for comparative phenological monitoring in an evergreen broad-leaved forest and a seasonal rain forest. Ecol. Inform. 2012, 10, 65–72. [Google Scholar] [CrossRef]
- Zhao, G.; Dong, J.; Cui, Y.; Liu, J.; Zhai, J.; He, T.; Zhou, Y.; Xiao, X. Evapotranspiration-dominated biogeophysical warming effect of urbanization in the Beijing-Tianjin-Hebei region, China. Clim. Dynam. 2019, 52, 1231–1245. [Google Scholar] [CrossRef]











| Site | Latitude | Longitude | Site-Years 1 |
|---|---|---|---|
| shiningrock | −82.774967 | 35.390159 | 14 |
| smokypurchase | −83.0733 | 35.586 | 14 |
| smokylook | −83.943113 | 35.632529 | 14 |
| mammothcave | −86.10194444 | 37.18583333 | 13 |
| nationalcapital | −77.069496 | 38.88818 | 13 |
| boundarywaters | −91.495506 | 47.946702 | 13 |
| caryinstitute | −73.7341 | 41.7839 | 11 |
| harvard | −72.1715 | 42.5378 | 11 |
| bartlettir | −71.2881 | 44.0646 | 11 |
| acadia | −68.26083333 | 44.37694444 | 11 |
| dollysods | −79.427041 | 39.099529 | 10 |
| morganmonroe | −86.4131 | 39.3231 | 10 |
| proctor | −72.866 | 44.525 | 10 |
| queens | −76.324 | 44.565 | 10 |
| umichbiological2 | −84.6976 | 45.5625 | 10 |
| joycekilmer | −83.795 | 35.257 | 9 |
| howland2 | −68.7418 | 45.2128 | 9 |
| umichbiological | −84.71382 | 45.55984 | 9 |
| woodshole | −70.6432 | 41.5495 | 8 |
| hubbardbrook | −71.701 | 43.9438 | 8 |
| snakerivermn | −93.24467 | 46.120556 | 8 |
| drippingsprings | −116.8 | 33.3 | 7 |
| oakridge2 | −84.3323 | 35.9311 | 7 |
| tonzi | −120.9658861 | 38.43091667 | 7 |
| missouriozarks | −92.2 | 38.7441 | 7 |
| northattleboroma | −71.31056 | 41.98369 | 7 |
| bostoncommon | −71.064145 | 42.355912 | 7 |
| readingma | −71.1272 | 42.5304 | 7 |
| harvardlph | −72.185 | 42.542 | 7 |
| turkeypointdbf | −80.5576 | 42.6353 | 7 |
| willowcreek | −90.07912 | 45.805986 | 7 |
| coweeta | −83.4275 | 35.0592 | 6 |
| alligatorriver | −75.9038 | 35.7879 | 6 |
| springfieldma | −72.585972 | 42.135162 | 6 |
| ashburnham | −71.926 | 42.6029 | 6 |
| downerwoods | −87.88076 | 43.07938 | 6 |
| uwmfieldsta | −88.0229 | 43.38709 | 6 |
| hubbardbrooksfws | −71.7407 | 43.9269 | 6 |
| russellsage | −91.974322 | 32.456961 | 5 |
| dukehw | −79.100371 | 35.973583 | 5 |
| bullshoals | −93.06663 | 36.562833 | 5 |
| shalehillsczo | −77.9041 | 40.6658 | 5 |
| worcester | −71.8428 | 42.2697 | 5 |
| arbutuslake | −74.23322 | 43.98207 | 5 |
| laurentides | −74.0055 | 45.9881 | 5 |
| canadaOA | −106.19779 | 53.62889 | 5 |
| silaslittle | −74.596 | 39.9137 | 4 |
| bbc2 | −72.185 | 42.542 | 4 |
| sanford | −84.464482 | 42.726777 | 4 |
| lacclair | −71.66957778 | 46.95209167 | 4 |
| marcell | −93.46925 | 47.5139 | 4 |
| NEON.D02.SERC.DP1.00033 | −76.56001 | 38.89008 | 3 |
| bbc5 | −70.6432 | 41.5495 | 3 |
| bbc1 | −72.174359 | 42.53508 | 3 |
| tfforest | −70.9505 | 43.1086 | 3 |
| arbutuslakeinlet | −74.24527 | 43.99336 | 3 |
| bbc7 | −71.2881 | 44.0646 | 3 |
| NEON.D08.LENO.DP1.00033 | −88.16122 | 31.85388 | 2 |
| NEON.D08.DELA.DP1.00033 | −87.803877 | 32.541727 | 2 |
| NEON.D11.CLBJ.DP1.00033 | −97.57 | 33.40123 | 2 |
| NEON.D07.GRSM.DP1.00033 | −83.50195 | 35.68896 | 2 |
| NEON.D07.ORNL.DP1.00033 | −84.282588 | 35.964128 | 2 |
| asuhighlands | −81.7032 | 36.2076 | 2 |
| NEON.D02.SCBI.DP1.00033 | −78.139494 | 38.892925 | 2 |
| usgsreston | −77.3676 | 38.9471 | 2 |
| NEON.D02.BLAN.DP1.00033 | −78.041788 | 39.033698 | 2 |
| macleish | −72.6804 | 42.4484 | 2 |
| NEON.D01.HARV.DP1.00033 | −72.17265 | 42.536911 | 2 |
| NEON.D01.BART.DP1.00033 | −71.287375 | 44.063869 | 2 |
| willamettepoplar | −123.1823 | 44.1368 | 2 |
| NEON.D05.TREE.DP1.00033 | −89.58572 | 45.49373 | 2 |
| NEON.D05.UNDE.DP1.00033 | −89.537254 | 46.23391 | 2 |
| millhaft | −2.29883 | 52.800796 | 2 |
| donanapajarera | −6.4432 | 36.9962 | 1 |
| NEON.D07.MLBS.DP1.00033 | −80.524847 | 37.378314 | 1 |
| robinson2 | −83.1576 | 37.4671 | 1 |
| pace | −78.2739 | 37.9229 | 1 |
| columbiamissouri | −92.1997 | 38.7441 | 1 |
| greenridge1 | −78.4067 | 39.6905 | 1 |
| hubbardbrooknfws | −71.7762 | 42.958 | 1 |
| NEON.D05.STEI.DP1.00033 | −89.58637 | 45.50894 | 1 |
| Bands | Landsat-7 | Bands | Sentinel-2 |
|---|---|---|---|
| Blue | Landsat8(480 nm) 1 = 0.0003 + 0.8474 × Landsat7(485 nm) 1 | Blue | Landsat8(480 nm) 1 = 0.0003 + 0.9570 × Sentinel2(490 nm) 1 |
| Red | Landsat8(655 nm) 1 = 0.0061 + 0.9047 × Landsat7(660 nm) 1 | Red | Landsat8(655 nm) 1 = 0.0041 + 0.9533 × Sentinel2(665 nm) 1 |
| NIR | Landsat8(865 nm) 1 = 0.0412 + 0.8462 × Landsat7(835 nm) 1 | NIR (Band 8A) | Landsat8(865 nm) 1 = 0.0077 + 0.9644 × Sentinel2(865 nm) 1 |
| SWIR | Landsat8 (1610 nm) 1 = 0.0254 + 0.8937 × Landsat7(1650 nm) 1 | SWIR | Landsat8(1610 nm) 1 = 0.0034 + 0.9522 × Sentinel2(1610 nm) 1 |
| Buffer | Metric | PhenoCam_GCC | Landsat and Sentinel-2 | MODIS | ||||
|---|---|---|---|---|---|---|---|---|
| EVI | NDVI | NIRv | EVI | NDVI | NIRv | |||
| 30 m | SOS | 1 | 0.87 | 0.79 | 0.87 | 0.84 | 0.75 | 0.85 |
| EOS | 1 | 0.77 | 0.85 | 0.71 | 0.85 | 0.85 | 0.85 | |
| 60 m | SOS | 1 | 0.86 | 0.79 | 0.87 | 0.84 | 0.75 | 0.85 |
| EOS | 1 | 0.80 | 0.85 | 0.69 | 0.85 | 0.85 | 0.85 | |
| 90 m | SOS | 1 | 0.86 | 0.79 | 0.87 | 0.84 | 0.75 | 0.85 |
| EOS | 1 | 0.80 | 0.82 | 0.74 | 0.85 | 0.85 | 0.85 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, K.; Yang, J.; Dong, J.; Zhao, G.; Cui, Y. Comparing Different Spatial Resolutions and Indices for Retrieving Land Surface Phenology for Deciduous Broadleaf Forests. Remote Sens. 2023, 15, 2266. https://0-doi-org.brum.beds.ac.uk/10.3390/rs15092266
Cui K, Yang J, Dong J, Zhao G, Cui Y. Comparing Different Spatial Resolutions and Indices for Retrieving Land Surface Phenology for Deciduous Broadleaf Forests. Remote Sensing. 2023; 15(9):2266. https://0-doi-org.brum.beds.ac.uk/10.3390/rs15092266
Chicago/Turabian StyleCui, Kailong, Jilin Yang, Jinwei Dong, Guosong Zhao, and Yaoping Cui. 2023. "Comparing Different Spatial Resolutions and Indices for Retrieving Land Surface Phenology for Deciduous Broadleaf Forests" Remote Sensing 15, no. 9: 2266. https://0-doi-org.brum.beds.ac.uk/10.3390/rs15092266