Evaluation of Continuous VNIR-SWIR Spectra versus Narrowband Hyperspectral Indices to Discriminate the Invasive Acacia longifolia within a Mediterranean Dune Ecosystem
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Collection of Field Spectra
2.3. Pre-Processing of Field Spectra
2.4. Calculation of Vegetation Indices and Red Edge Parameters
2.5. Classification
3. Results
3.1. Separating Acacia longifolia from Adjacent Mediterranean Dune Vegetation at the Leaf and Canopy Level
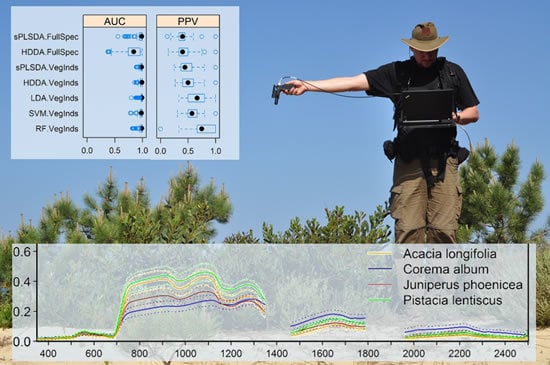
3.2. Comparison of Model Performance Using Full Spectrum Data and Hyperspectral Narrowband Vegetation Indices
3.3. Identification of the Optimal Algorithm to Distinguish Acacia longifolia from Adjacent Vegetation
3.4. Identification of the Most Important Variables
4. Discussion
4.1. Distinguishing Acacia longifolia from Adjacent Vegetation at the Leaf and Canopy Level
4.2. Identifying the Optimal Classifier
4.3. Interpretation of the Relevant Vegetation Indices for Identifying Acacia longifolia at the Canopy Level
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix

References
- Pyšek, P.; Jarošík, V.; Hulme, P.E.; Pergl, J.; Hejda, M.; Schaffner, U.; Vilà, M. A global assessment of invasive plant impacts on resident species, communities and ecosystems: The interaction of impact measures, invading species’ traits and environment. Glob. Change Biol. 2012, 18, 1725–1737. [Google Scholar] [CrossRef]
- Vilà, M.; Basnou, C.; Pyšek, P.; Josefsson, M.; Genovesi, P.; Gollasch, S.; Nentwig, W.; Olenin, S.; Roques, A.; Roy, D.; et al. How well do we understand the impacts of alien species on ecosystem services? A pan-European, cross-taxa assessment. Front. Ecol. Environ. 2010, 8, 135–144. [Google Scholar] [CrossRef] [Green Version]
- He, K.S.; Rocchini, D.; Neteler, M.; Nagendra, H. Benefits of hyperspectral remote sensing for tracking plant invasions: Plant invasion and hyperspectral remote sensing. Divers. Distrib. 2011, 17, 381–392. [Google Scholar] [CrossRef]
- Huang, C.; Asner, G.P. Applications of remote sensing to alien invasive plant studies. Sensors 2009, 9, 4869–4889. [Google Scholar] [CrossRef] [PubMed]
- Rocchini, D.; Andreo, V.; Förster, M.; Garzon-Lopez, C.X.; Gutierrez, A.P.; Gillespie, T.W.; Hauffe, H.C.; He, K.S.; Kleinschmit, B.; Mairota, P.; et al. Potential of remote sensing to predict species invasions: A modelling perspective. Prog. Phys. Geogr. 2015, 39, 283–309. [Google Scholar] [CrossRef]
- Asner, G.P.; Wessman, C.A.; Bateson, C.; Privette, J.L. Impact of tissue, canopy, and landscape factors on the hyperspectral reflectance variability of arid ecosystems. Remote Sens. Environ. 2000, 74, 69–84. [Google Scholar] [CrossRef]
- Clark, M.; Roberts, D.; Clark, D. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A. Species-Level differences in hyperspectral metrics among tropical rainforest trees as determined by a tree-based classifier. Remote Sens. 2012, 4, 1820–1855. [Google Scholar] [CrossRef]
- Féret, J.-B.; Asner, G.P. Spectroscopic classification of tropical forest species using radiative transfer modeling. Remote Sens. Environ. 2011, 115, 2415–2422. [Google Scholar] [CrossRef]
- Kalacska, M.; Bohlman, S.; Sanchez-Azofeifa, G.A.; Castro-Esau, K.; Caelli, T. Hyperspectral discrimination of tropical dry forest lianas and trees: Comparative data reduction approaches at the leaf and canopy levels. Remote Sens. Environ. 2007, 109, 406–415. [Google Scholar] [CrossRef]
- Roberts, D.A.; Ustin, S.L.; Ogunjemiyo, S.; Greenberg, J.; Dobrowski, S.Z.; Chen, J.; Hinckley, T.M. Spectral and structural measures of northwest forest vegetation at leaf to landscape scales. Ecosystems 2004, 7, 545–562. [Google Scholar] [CrossRef]
- Burkholder, A.; Warner, T.A.; Culp, M.; Landenberger, R. Seasonal trends in separability of leaf reflectance spectra for Ailanthus altissima and four other tree species. Photogramm. Eng. Remote Sens. 2011, 77, 793–804. [Google Scholar] [CrossRef]
- Taylor, S.L.; Hill, R.A.; Edwards, C. Characterising invasive non-native Rhododendron ponticum spectra signatures with spectroradiometry in the laboratory and field: Potential for remote mapping. ISPRS J. Photogramm. Remote Sens. 2013, 81, 70–81. [Google Scholar] [CrossRef]
- Fernandes, M.R.; Aguiar, F.C.; Silva, J.M.N.; Ferreira, M.T.; Pereira, J.M.C. Spectral discrimination of giant reed (Arundo donax L.): A seasonal study in riparian areas. ISPRS J. Photogramm. Remote Sens. 2013, 80, 80–90. [Google Scholar] [CrossRef]
- Ouyang, Z.-T.; Gao, Y.; Xie, X.; Guo, H.-Q.; Zhang, T.-T.; Zhao, B. Spectral discrimination of the invasive plant Spartina alterniflora at multiple phenological stages in a saltmarsh wetland. PLoS ONE 2013, 8, e67315. [Google Scholar] [CrossRef] [PubMed]
- Fassnacht, F.E.; Neumann, C.; Forster, M.; Buddenbaum, H.; Ghosh, A.; Clasen, A.; Joshi, P.K.; Koch, B. Comparison of feature reduction algorithms for classifying tree species with hyperspectral data on three central European test sites. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2547–2561. [Google Scholar] [CrossRef]
- Somers, B.; Asner, G.P. Multi-temporal hyperspectral mixture analysis and feature selection for invasive species mapping in rainforests. Remote Sens. Environ. 2013, 136, 14–27. [Google Scholar] [CrossRef]
- Tits, L.; De Keersmaecker, W.; Somers, B.; Asner, G.P.; Farifteh, J.; Coppin, P. Hyperspectral shape-based unmixing to improve intra- and interclass variability for forest and agro-ecosystem monitoring. ISPRS J. Photogramm. Remote Sens. 2012, 74, 163–174. [Google Scholar] [CrossRef]
- Prospere, K.; McLaren, K.; Wilson, B. Plant species discrimination in a tropical wetland using in situ hyperspectral data. Remote Sens. 2014, 6, 8494–8523. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Mariotto, I.; Gumma, M.K.; Middleton, E.M.; Landis, D.R.; Huemmrich, K.F. Selection of hyperspectral narrowbands (HNBs) and composition of hyperspectral twoband vegetation indices (HVIs) for biophysical characterization and discrimination of crop types using field reflectance and Hyperion/EO-1 Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 427–439. [Google Scholar] [CrossRef]
- Castro-Esau, K. Discrimination of lianas and trees with leaf-level hyperspectral data. Remote Sens. Environ. 2004, 90, 353–372. [Google Scholar] [CrossRef]
- Hostert, P.; Swayne, F.; Cohen, W.B.; Chipman, J. The role of remote sensing in LTER Projects. In Long-Term Ecological Research; Müller, F., Baessler, C., Schubert, H., Klotz, S., Eds.; Springer: Dordrecht, NL, USA, 2010; pp. 131–142. [Google Scholar]
- Jiménez, M.; Díaz-Delgado, R. Towards a standard plant species spectral library protocol for vegetation mapping: A case study in the shrubland of Doñana National Park. ISPRS Int. J. Geo-Inf. 2015, 4, 2472–2495. [Google Scholar] [CrossRef]
- Ghosh, A.; Joshi, P.K. A comparison of selected classification algorithms for mapping bamboo patches in lower Gangetic plains using very high resolution WorldView 2 imagery. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 298–311. [Google Scholar] [CrossRef]
- Vaiphasa, C.; Skidmore, A.K.; de Boer, W.F.; Vaiphasa, T. A hyperspectral band selector for plant species discrimination. ISPRS J. Photogramm. Remote Sens. 2007, 62, 225–235. [Google Scholar] [CrossRef]
- Heim, R.; Jürgens, N.; Große-Stoltenberg, A.; Oldeland, J. The effect of epidermal structures on leaf spectral signatures of ice plants (Aizoaceae). Remote Sens. 2015, 7, 16901–16914. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Vescovo, L.; Gianelle, D. The role of spectral resolution and classifier complexity in the analysis of hyperspectral images of forest areas. Remote Sens. Environ. 2009, 113, 2345–2355. [Google Scholar] [CrossRef]
- Bouveyron, C.; Girard, S.; Schmid, C. High-Dimensional discriminant analysis. Commun. Stat. Theory Methods 2007, 36, 2607–2623. [Google Scholar] [CrossRef]
- Richardson, D.M.; Carruthers, J.; Hui, C.; Impson, F.A.C.; Miller, J.T.; Robertson, M.P.; Rouget, M.; Le Roux, J.J.; Wilson, J.R.U. Human-mediated introductions of Australian acacias—A global experiment in biogeography: Wattles: A model group for invasion science. Divers. Distrib. 2011, 17, 771–787. [Google Scholar] [CrossRef]
- Hellmann, C.; Sutter, R.; Rascher, K.G.; Máguas, C.; Correia, O.; Werner, C. Impact of an exotic N2-fixing Acacia on composition and N status of a native Mediterranean community. Acta Oecol. 2011, 37, 43–50. [Google Scholar] [CrossRef]
- Marchante, E.; Kjøller, A.; Struwe, S.; Freitas, H. Short- and long-term impacts of Acacia longifolia invasion on the belowground processes of a Mediterranean coastal dune ecosystem. Appl. Soil Ecol. 2008, 40, 210–217. [Google Scholar] [CrossRef]
- Marchante, H.; Marchante, E.; Freitas, H.; Hoffmann, J.H. Temporal changes in the impacts on plant communities of an invasive alien tree, Acacia longifolia. Plant Ecol. 2015, 216, 1481–1498. [Google Scholar] [CrossRef]
- Rascher, K.G.; Große-Stoltenberg, A.; Máguas, C.; Meira-Neto, J.A.A.; Werner, C. Acacia longifolia invasion impacts vegetation structure and regeneration dynamics in open dunes and pine forests. Biol. Invasions 2011, 13, 1099–1113. [Google Scholar] [CrossRef]
- Calviño-Cancela, M.; Méndez-Rial, R.; Reguera-Salgado, J.; Martín-Herrero, J. Alien plant monitoring with ultralight airborne imaging spectroscopy. PLoS ONE 2014, 9, e102381. [Google Scholar]
- Sociedade Portuguesa de Botânica. Flora-On: Flora de Portugal Interactiva. Available online: http://www.flora-on.pt/ (accessed on 27 November 2015).
- Guedes, A.; Ribeiro, H.; Valentim, B.; Rodrigues, A.; Sant’Ovaia, H.; Abreu, I.; Noronha, F. Characterization of soils from the Algarve region (Portugal): A multidisciplinary approach for forensic applications. Sci. Justice 2011, 51, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Global Invasive Species Database. Distribution of Acacia longifolia. Available online: http://www.issg.org/database/species/distribution.asp?si=1662&fr=1&sts=sss&lang=EN (accessed on 19 January 2016).
- Richardson, D.M.; Le Roux, J.J.; Wilson, J.R. Australian acacias as invasive species: Lessons to be learnt from regions with long planting histories. South. For. J. For. Sci. 2015, 77, 31–39. [Google Scholar] [CrossRef]
- Instituto Hórus de Desenvolvimento e Conservação Ambiental. Base de Dados Nacional de Espécies Exóticas Invasoras I3N Brasil. Available online: http://i3n.institutohorus.org.br/www (accessed on 19 January 2016).
- Alberio, C.; Comparatore, V. Patterns of woody plant invasion in an Argentinean coastal grassland. Acta Oecol. 2014, 54, 65–71. [Google Scholar] [CrossRef]
- Werner, C.; Peperkorn, R.; Máguas, C.; Beyschlag, W. Competitive balance between the alien invasive Acacia longifolia and native Mediterranean species. In Plant Invasions: Human perception, Ecological Impacts and Management; Tokarska-Guzik, B., Brock, J.H., Brundu, G., Child, L., Daehler, C.C., Pyšek, P., Eds.; Backhuys Publishers: Leiden, The Netherlands, 2008; pp. 261–275. [Google Scholar]
- Peperkorn, R.; Werner, C.; Beyschlag, W. Phenotypic plasticity of an invasive acacia versus two native Mediterranean species. Funct. Plant Biol. 2005, 32, 933–944. [Google Scholar] [CrossRef]
- Marchante, H.; Marchante, E.; Freitas, H. Invasion of the Portuguese dune ecosystems by the exotic species Acacia longifolia (Andrews) Willd: Effects at the community level. In Plant Invasions: Ecological Threats and Management Solutions; Child, L.E., Brock, J.H., Brundu, G., Prach, K., Pyšek, P., Wade, P., Williamson, M., Eds.; Backhuys Publishers: Leiden, The Netherlands, 2003; pp. 75–85. [Google Scholar]
- Rascher, K.G.; Hellmann, C.; Máguas, C.; Werner, C. Community scale 15N isoscapes: Tracing the spatial impact of an exotic N2-fixing invader: Community scale isoscapes. Ecol. Lett. 2012, 15, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Rascher, K.G.; Große-Stoltenberg, A.; Máguas, C.; Werner, C. Understory invasion by Acacia longifolia alters the water balance and carbon gain of a Mediterranean pine forest. Ecosystems 2011, 14, 904–919. [Google Scholar] [CrossRef]
- Marchante, H.; Freitas, H.; Hoffmann, J.H. The potential role of seed banks in the recovery of dune ecosystems after removal of invasive plant species: The potential role of seed-banks in the recovery of dune ecosystems. Appl. Veg. Sci. 2011, 14, 107–119. [Google Scholar] [CrossRef]
- Instituto da Conservação da Natureza e da Biodiversidade. Sítio Comporta/Galé; Plano Sectorial da Rede Natura 2000; Instituto da Conservação da Natureza e da Biodiversidade: Lisbon, Portugal, 2005. [Google Scholar]
- European Topic Centre on Biological Diversity. Mediterranean Terrestrial Region. Reference List; European Topic Centre on Biological Diversity: Paris, France, 2015. [Google Scholar]
- Pinto, M.J. Guia das Plantas e dos Ecossistemas da Reserva Natural das Lagoas de Santo André e da Sancha; Instituto da Conservação da Natureza e das Florestas (ICNF): Lisbon, Portugal, 2014. [Google Scholar]
- Hellmann, C.; Große-Stoltenberg, A.; Lauströer, V.; Oldeland, J.; Werner, C. Retrieving nitrogen isotopic signatures from fresh leaf reflectance spectra: Disentangling δ15N from biochemical and structural leaf properties. Front. Plant Sci. 2015, 6, 307. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.; Große-Stoltenberg, A.; Römer, M.; Oldeland, J. Field spectroscopy in the VNIR-SWIR region to discriminate between Mediterranean native plants and exotic-invasive shrubs based on leaf tannin content. Remote Sens. 2015, 7, 1225–1241. [Google Scholar] [CrossRef] [Green Version]
- ASD Inc. FieldSpec4 User Manual; ASD Inc., a PANalytical Company: Boulder, CO, USA, 2015. [Google Scholar]
- Foley, S.; Rivard, B.; Sanchezazofeifa, G.; Calvo, J. Foliar spectral properties following leaf clipping and implications for handling techniques. Remote Sens. Environ. 2006, 103, 265–275. [Google Scholar] [CrossRef]
- Pimstein, A.; Notesco, G.; Ben-Dor, E. Performance of three identical spectrometers in retrieving soil reflectance under laboratory conditions. Soil Sci. Soc. Am. J. 2011, 75, 746–759. [Google Scholar] [CrossRef]
- Máguas, C.; Rascher, K.G.; Martins-Loução, A.; Carvalho, P.; Pinho, P.; Ramos, M.; Correia, O.; Werner, C. Responses of woody species to spatial and temporal ground water changes in coastal sand dune systems. Biogeosciences 2011, 8, 3823–3832. [Google Scholar] [CrossRef]
- De Jong, S.M.; Addink, E.A.; Hoogenboom, P.; Nijland, W. The spectral response of Buxus sempervirens to different types of environmental stress—A laboratory experiment. ISPRS J. Photogramm. Remote Sens. 2012, 74, 56–65. [Google Scholar] [CrossRef]
- Liew, O.W.; Chong, P.C.J.; Li, B.; Asundi, A.K. Signature optical cues: Emerging technologies for monitoring plant health. Sensors 2008, 8, 3205–3239. [Google Scholar] [CrossRef]
- Krawczyk, B.; Woźniak, M.; Herrera, F. On the usefulness of one-class classifier ensembles for decomposition of multi-class problems. Pattern Recognit. 2015, 48, 3969–3982. [Google Scholar] [CrossRef]
- Rifkin, R.; Klautau, A. In defense of one-vs-all classification. J. Mach. Learn. Res. 2004, 5, 101–141. [Google Scholar]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Anthony, G.; Gregg, H.; Tshilidzi, M. Image classification using SVMs: One-against-one vs. one-against-all. In Proceedings of the 28th Asian Conference on Remote Sensing, Kuala Lumpur, Malaysia, 12–16 November 2007.
- Lawrence, R.L.; Wood, S.D.; Sheley, R.L. Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (randomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Instituto da Conservação da Natureza e das Florestas (ICNF). Plano Setorial—Flora; Fichas de caracterização e gestão das espécies constantes no Anexo II da Diretiva Habitats-Flora. Available online: http://www.icnf.pt/portal/naturaclas/rn2000/p-set/psrn-flora (accessed on 27 November 2015).
- Dorigo, W.; Bachmann, M.; Heldens, W. AS Toolbox & Processing of Field Spectra: User’s Manual; German Aerospace Center (DLR): Oberpfaffenhofen, Germany, 2006. [Google Scholar]
- Lehnert, L.W.; Meyer, H.; Bendix, J. hsdar: Manage, Analyse and Simulate Hyperspectral Data in R. R Package Version 0.3.1. Available online: https://cran.r-project.org/web/packages/hsdar (accessed on 8 October 2015).
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013. [Google Scholar]
- Chung, D.; Keles, S. Sparse partial least squares classification for high dimensional data. Stat. Appl. Genet. Mol. Biol. 2010, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Berge, L.; Bouveyron, C.; Girard, S. HDclassif: An R package for model-based clustering and discriminant analysis of high-dimensional data. J. Stat. Softw. 2012, 46, 1–29. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Pu, R. Broadleaf species recognition with in situ hyperspectral data. Int. J. Remote Sens. 2009, 30, 2759–2779. [Google Scholar] [CrossRef]
- Fernández-Delgado, M.; Cernadas, E.; Barro, S.; Amorim, D. Do we need hundreds of classifiers to solve real world classification problems? J. Mach. Learn. Res. 2014, 15, 3133–3181. [Google Scholar]
- van der Linden, S.; Rabe, A.; Held, M.; Jakimow, B.; Leitão, P.; Okujeni, A.; Schwieder, M.; Suess, S.; Hostert, P. The EnMAP-Box—A toolbox and application programming interface for EnMAP data processing. Remote Sens. 2015, 7, 11249–11266. [Google Scholar] [CrossRef]
- Chung, D.; Chun, H.; Keles, S. spls: Sparse Partial Least Squares (SPLS) Regression and Classification. R package version 2.2-1. Available online: https://cran.r-project.org/web/packages/spls (accessed on 30 November 2015).
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Karatzoglou, A.; Smola, A.; Hornik, K. The Kernlab Package. Kernel-Based Machine Learning Lab. R package version 0.9.-22. Available online: https://cran.r-project.org/web/packages/kernlab (accessed on 4 November 2015).
- Japkowicz, N. Concept-learning in the presence of between-class and within-class imbalances. In Advances in Artificial Intelligence; Springer: Berlin, Germany, 2001; pp. 67–77. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: Display and Analyze ROC Curves. R Package Version 1.8. Available online: https://cran.r-project.org/web/packages/pROC (accessed on 30 November 2015).
- Kuhn, M. caret: Classification and Regression Training. R package version 6.0-52. Available online: https://cran.r-project.org/web/packages/caret (accessed on 29 November 2015).
- Nagler, P.L.; Inoue, Y.; Glenn, E.P.; Russ, A.L.; Daughtry, C.S.T. Cellulose absorption index (CAI) to quantify mixed soil–plant litter scenes. Remote Sens. Environ. 2003, 87, 310–325. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Pushnik, J.C.; Dobrowski, S.; Ustin, S.L. Steady-state chlorophyll a fluorescence detection from canopy derivative reflectance and double-peak red-edge effects. Remote Sens. Environ. 2003, 84, 283–294. [Google Scholar] [CrossRef]
- Apan, A.; Held, A.; Phinn, S.; Markley, J. Detecting sugarcane “orange rust” disease using EO-1 Hyperion hyperspectral imagery. Int. J. Remote Sens. 2004, 25, 489–498. [Google Scholar] [CrossRef] [Green Version]
- Huete, A.; Lui, H.Q.; Batchily, K.; van Leeuwen, W. A comparison of vegetation indices over a global set of TM images for EOS-MODIS. Remote Sens. Environ. 1997, 59, 440–451. [Google Scholar] [CrossRef]
- Galvão, L.S.; Formaggio, A.R.; Tisot, D.A. Discrimination of sugarcane varieties in Southeastern Brazil with EO-1 Hyperion data. Remote Sens. Environ. 2005, 94, 523–534. [Google Scholar] [CrossRef]
- Lobell, D.B.; Asner, G.P. Subpixel canopy cover estimation of coniferous forests in Oregon using SWIR imaging spectrometry. J. Geophys. Res. Atmospheres 2001, 106, 5151–5160. [Google Scholar]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Datt, B. Remote sensing of water content in Eucalyptus leaves. Aust. J. Bot. 1999, 47, 909–923. [Google Scholar] [CrossRef]
- Penuelas, J.; Gamon, J.A.; Fredeen, A.L.; Merino, J.; Field, C.B. Reflectance indices associated with physiological changes in nitrogen- and water-limited sunflower leaves. Remote Sens. Environ. 1994, 48, 135–146. [Google Scholar] [CrossRef]
- Bach, H. Die Bestimmung Hydrologischer und Landwirtschaftlicher Oberflächenparameter aus Hyperspektralen Fernerkundungsdaten; Münchener Geographische Abhandlungen: München, Germany, 1995. [Google Scholar]
- Lichtenthaler, H.K.; Lang, M.; Sowinska, M.; Heisel, F.; Miehé, J.A. Detection of vegetation stress via a new high resolution fluorescence imaging system. J. Plant Physiol. 1996, 148, 599–612. [Google Scholar] [CrossRef]
- Datt, B. Remote sensing of chlorophyll a, chlorophyll b, chlorophyll a+b, and total carotenoid content in eucalyptus leaves. Remote Sens. Environ. 1998, 66, 111–121. [Google Scholar] [CrossRef]
- Le Maire, G.; Francois, C.; Soudani, K.; Berveiller, D.; Pontailler, J.; Breda, N.; Genet, H.; Davi, H.; Dufrene, E. Calibration and validation of hyperspectral indices for the estimation of broadleaved forest leaf chlorophyll content, leaf mass per area, leaf area index and leaf canopy biomass. Remote Sens. Environ. 2008, 112, 3846–3864. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Serrano, L.; Penuelas, J.; Ustin, S.L. Remote sensing of nitrogen and lignin in Mediterranean vegetation from AVIRIS data: Decomposing biochemical from structural signals. Remote Sens. Environ. 2002, 81, 355–364. [Google Scholar] [CrossRef]
- Garrity, S.R.; Eitel, J.U.H.; Vierling, L.A. Disentangling the relationships between plant pigments and the photochemical reflectance index reveals a new approach for remote estimation of carotenoid content. Remote Sens. Environ. 2011, 115, 628–635. [Google Scholar] [CrossRef]
- Merzlyak, M.D.; Gitelson, A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 135–141. [Google Scholar] [CrossRef]
- Penuelas, J.; Baret, F.; Filella, I. Semiempirical indexes to assess carotenoids chlorophyll-a ratio from leaf spectral reflectance. Photosynthetica 1995, 31, 220–231. [Google Scholar]
- Le Maitre, D.C.; Gaertner, M.; Marchante, E.; Ens, E.-J.; Holmes, P.M.; Pauchard, A.; O’Farrell, P.J.; Rogers, A.M.; Blanchard, R.; Blignaut, J.; Richardson, D.M. Impacts of invasive Australian acacias: Implications for management and restoration: Australian acacias: Linking impacts and restoration. Divers. Distrib. 2011, 17, 1015–1029. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Teluguntla, P.; Gumma, M.K.; Dheeravath, V. Hyperspectral remote sensing for terrestrial applications. In Remote Sensing Handbook: Land Resources Monitoring, Modeling, and Mapping with Remote Sensing; Thenkabail, P.S., Ed.; CRC Press: Boca Raton, FL, USA, 2015; pp. 201–233. [Google Scholar]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Advances in hyperspectral remote sensing of vegetation and agricultural croplands. In Hyperspectral Remote Sensing of Vegetation; Thenkabail, P.S., Lyon, J.G., Huete, A., Eds.; CRC Press: Boca Raton, FL, USA, 2011; pp. 3–36. [Google Scholar]
- Millard, K.; Richardson, M. On the importance of training data sample selection in random forest image classification: A case study in peatland ecosystem mapping. Remote Sens. 2015, 7, 8489–8515. [Google Scholar] [CrossRef]
- Pal, M.; Foody, G.M. Feature selection for classification of hyperspectral data by SVM. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2297–2307. [Google Scholar] [CrossRef]
- Cánovas-García, F.; Alonso-Sarría, F. Optimal combination of classification algorithms and feature ranking methods for object-based classification of submeter resolution Z/I-Imaging DMC imagery. Remote Sens. 2015, 7, 4651–4677. [Google Scholar] [CrossRef]
- Bandos, T.V.; Bruzzone, L.; Camps-Valls, G. Classification of hyperspectral images with regularized linear discriminant analysis. IEEE Trans. Geosci. Remote Sens. 2009, 47, 862–873. [Google Scholar] [CrossRef]
- Hamada, Y.; Stow, D.A.; Coulter, L.L.; Jafolla, J.C.; Hendricks, L.W. Detecting Tamarisk species (Tamarix spp.) in riparian habitats of Southern California using high spatial resolution hyperspectral imagery. Remote Sens. Environ. 2007, 109, 237–248. [Google Scholar] [CrossRef]
- Oldeland, J.; Dorigo, W.; Wesuls, D.; Jürgens, N. Mapping bush encroaching species by seasonal differences in hyperspectral imagery. Remote Sens. 2010, 2, 1416–1438. [Google Scholar] [CrossRef]
- Godoy, O.; de Lemos-Filho, J.P.; Valladares, F. Invasive species can handle higher leaf temperature under water stress than Mediterranean natives. Environ. Exp. Bot. 2011, 71, 207–214. [Google Scholar] [CrossRef]
- Australian Biological Resources Study (ABRS). Flora of Australia Online. In Acacia longifolia. Available online: http://www.anbg.gov.au/abrs/online-resources/flora/stddisplay.xsql?pnid=41939 (accessed on 30 November 2015).
- Escribano, P.; Palacios-Orueta, A.; Oyonarte, C.; Chabrillat, S. Spectral properties and sources of variability of ecosystem components in a Mediterranean semiarid environment. J. Arid Environ. 2010, 74, 1041–1051. [Google Scholar] [CrossRef]




| Species | Leaf | Canopy | Growth Form | Family | Status |
|---|---|---|---|---|---|
| Acacia longifolia | 74 | 18 | P | Fabaceae | inv |
| Acacia saligna | 24 | 16 | P | Fabaceae | inv |
| Armeria pungens | 5 | 7 | C | Plumbaginaceae | |
| Arundo donax | 8 | n.a. | Ph | Poaceae | inv |
| Calluna vulgaris | 8 | 1 | P, C | Ericaceae | |
| Carpobrotus edulis | 11 | 22 | C | Aizoaceae | inv |
| Cistus ladanifer | n.a. | 5 | P | Cistaceae | |
| Cistus salviifolius | 12 | 16 | P, C | Cistaceae | |
| Corema album | 120 | 23 | P | Ericaceae | |
| Daphne gnidium | 3 | 1 | C, P | Thymelaeaceae | |
| Halimium calycinum | 21 | 16 | P | Cistaceae | |
| Halimium halimifolium | 49 | 14 | C, P | Cistaceae | |
| Helichrysum italicum subsp. picardii | 13 | 16 | C | Asteraceae | |
| Juniperus navicularis | 10 | 10 | P, C | Cupressaceae | end |
| Juniperus phoenicea subsps. turbinata | 34 | 23 | P, C | Cupressaceae | |
| Lavandula stoechas | 20 | 14 | P, C | Lamiaceae | |
| Pancratium maritimum | 9 | n.a. | G | Amaryllidaceae | |
| Paraserianthes lophantha | 16 | n.a. | P | Fabaceae | inv |
| Phillyrea angustifolia | 3 | 3 | P | Oleaceae | |
| Pinus pinaster | 42 | 17 | P | Pinaceae | |
| Pistacia lentiscus | 43 | 24 | P | Anacardiaceae | |
| Rosmarinus officinalis | 29 | 24 | C, P | Lamiaceae | |
| Santolina impressa | 30 | 19 | C | Asteraceae | end, AnnII |
| Sedum sediforme | 1 | 1 | C | Crassulaceae | |
| Stauracanthus sp. | n.a. | 20 | P | Fabaceae | |
| Thymus camphoratus | 7 | 8 | C | Lamiaceae | end, AnnII |
| Thymus carnosus | 15 | 19 | C | Lamiaceae | end, AnnII |
| Ulex australis | n.a. | 9 | P, C | Fabaceae | end |
| Class | Leaf | Canopy | ||
|---|---|---|---|---|
| Training | Test | Training | Test | |
| Acacia longifolia | 56 | 18 | 14 | 4 |
| Other plant species | 400 | 133 | 246 | 82 |
| Vegetation Index | Related to | Sensor Level | Formula | References | |
|---|---|---|---|---|---|
| CAI | Cellulose Absorption Index | Cellulose | L *, C * | [83] | |
| DPI | Double Peak Index | Stress | L *, C * | [84] | |
| DWSI2 | Disease-Water Stress Index 2 | Disease-Water Stress | L *, C | [85] | |
| EVI | Enhanced Vegetation Index | Biomass/LAI | L *, C | [86] | |
| LWVI1 | Leaf Water Vegetation Index 1 | Water | L *, C * | [87] | |
| SWIR.VI | Shortwave-Infrared Vegetation Index | Vegetation Cover | L *, C * | [88] | |
| TCARI2 | Transformed Chlorophyll Absorption Ratio Index, bands used as in [89] | Chlorophyll | L *, C * | [89] | |
| Datt7 | n.a. | Water | C * | [90] | |
| DWSI3 | Disease-Water Stress Index 3 | Disease-Water Stress | C | [85] | |
| EGFR | Edge-Green First Derivative Ratio | Chlorophyll, Nitrogen | C * | [91] | |
| I0 | wavelength of the min. reflectance of the red-edge | Biomass | C | [92] | |
| SR7 | Fluorescence Ratio Blue/Red | Fluorescence, Stress | C | [93] | |
| Datt4 | n.a. | Pigments | L * | [94] | |
| Datt8 | n.a. | Water | L * | [90] | |
| DDn | new Double Difference Index | Chlorophyll | L * | [95] | |
| mSR | modified Simple Ratio | Chlorophyll | L * | [96] | |
| NDLI | Normalized Difference Lignin Index | Lignin | L * | [97] | |
| NDNI | Normalized Difference Nitrogen Index | Nitrogen | L * | [97] | |
| PRI.CI2 | Photochemical Reflection Index x Chlorophyll Content | Carotenoid | L * | [98] | |
| PSRI | Plant Senescing Reflectance Index | Leaf Senescence | L * | [99] | |
| SRPI | Simple Ratio Pigment Index | Pigments | L * | [100] | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Große-Stoltenberg, A.; Hellmann, C.; Werner, C.; Oldeland, J.; Thiele, J. Evaluation of Continuous VNIR-SWIR Spectra versus Narrowband Hyperspectral Indices to Discriminate the Invasive Acacia longifolia within a Mediterranean Dune Ecosystem. Remote Sens. 2016, 8, 334. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8040334
Große-Stoltenberg A, Hellmann C, Werner C, Oldeland J, Thiele J. Evaluation of Continuous VNIR-SWIR Spectra versus Narrowband Hyperspectral Indices to Discriminate the Invasive Acacia longifolia within a Mediterranean Dune Ecosystem. Remote Sensing. 2016; 8(4):334. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8040334
Chicago/Turabian StyleGroße-Stoltenberg, André, Christine Hellmann, Christiane Werner, Jens Oldeland, and Jan Thiele. 2016. "Evaluation of Continuous VNIR-SWIR Spectra versus Narrowband Hyperspectral Indices to Discriminate the Invasive Acacia longifolia within a Mediterranean Dune Ecosystem" Remote Sensing 8, no. 4: 334. https://0-doi-org.brum.beds.ac.uk/10.3390/rs8040334