The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation
Abstract
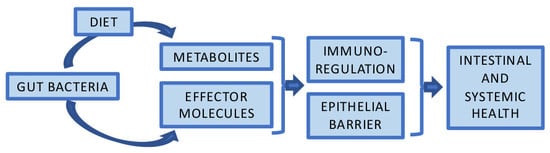
:1. Introduction
2. Intestinal Barrier Function
3. Proteobacteria as a Marker of Dysbiosis and Mediator of Inflammation
4. Health-Associated Commensal Bacteria
4.1. Bifidobacterium spp.
4.2. Akkermansia muciniphila
4.3. Faecalibacterium prausnitzii
4.4. Roseburia intestinalis
4.5. Eubacterium hallii
4.6. Bacteroides spp.
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Rajilic-Stojanovic, M.; de Vos, W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef] [PubMed]
- Salonen, A.; Salojarvi, J.; Lahti, L.; de Vos, W.M. The adult intestinal core microbiota is determined by analysis depth and health status. Clin. Microbiol. Infect. 2012, 18 (Suppl. 4), 16–20. [Google Scholar] [CrossRef] [PubMed]
- Ouwerkerk, J.P.; de Vos, W.M.; Belzer, C. Glycobiome: Bacteria and mucus at the epithelial interface. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Jarnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854. [Google Scholar] [CrossRef] [PubMed]
- Nava, G.M.; Friedrichsen, H.J.; Stappenbeck, T.S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011, 5, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Von Hertzen, L.; Beutler, B.; Bienenstock, J.; Blaser, M.; Cani, P.D.; Eriksson, J.; Farkkila, M.; Haahtela, T.; Hanski, I.; Jenmalm, M.C.; et al. Helsinki alert of biodiversity and health. Ann. Med. 2015, 47, 218–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, B.O.; Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Satokari, R. Contentious host-microbiota relationship in inflammatory bowel disease—Can foes become friends again? Scand. J. Gastroenterol. 2015, 50, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Lawley, T.D. Therapeutic modulation of intestinal dysbiosis. Pharmacol. Res. 2013, 69, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Rigottier-Gois, L. Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J. 2013, 7, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.C.; Wang, J.T.; Wei, S.C.; Ni, Y.H. Host-microbial interactions and regulation of intestinal epithelial barrier function: From physiology to pathology. World J. Gastrointest. Pathophysiol. 2012, 3, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Muniz, L.R.; Knosp, C.; Yeretssian, G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front. Immunol. 2012, 3, 310. [Google Scholar] [CrossRef] [PubMed]
- Goll, R.; van Beelen Granlund, A. Intestinal barrier homeostasis in inflammatory bowel disease. Scand. J. Gastroenterol. 2015, 50, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H. Intestinal M cells. J. Biochem. 2016, 159, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Ladinsky, M.S.; Yu, K.B.; Sanders, J.G.; Yoo, B.B.; Chou, W.C.; Conner, M.E.; Earl, A.M.; Knight, R.; Bjorkman, P.J.; et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018, 360, 795–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulluwishewa, D.; Anderson, R.C.; McNabb, W.C.; Moughan, P.J.; Wells, J.M.; Roy, N.C. Regulation of tight junction permeability by intestinal bacteria and dietary components. J. Nutr. 2011, 141, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 2013, 70, 631–659. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Lewis, P.; Samuelson, D.; Liboni, K.; Neu, J. Glutamine regulates Caco-2 cell tight junction proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 287, G726–G733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, Y.Y.; Ha, C.W.; Hoffmann, J.M.; Oscarsson, J.; Dinudom, A.; Mather, T.J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Effects of dietary fat profile on gut permeability and microbiota and their relationships with metabolic changes in mice. Obesity 2015, 23, 1429–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [PubMed]
- Heumann, D.; Roger, T. Initial responses to endotoxins and Gram-negative bacteria. Clin. Chim. Acta 2002, 323, 59–72. [Google Scholar] [CrossRef]
- Cochet, F.; Peri, F. The Role of Carbohydrates in the Lipopolysaccharide (LPS)/Toll-Like Receptor 4 (TLR4) Signalling. Int. J. Mol. Sci. 2017, 18, 2318. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Normark, S.; Schweda, E.K.; Oscarson, S.; Richter-Dahlfors, A. Structural requirements for TLR4-mediated LPS signalling: A biological role for LPS modifications. Microbes Infect. 2003, 5, 1057–1063. [Google Scholar] [CrossRef]
- Teghanemt, A.; Zhang, D.; Levis, E.N.; Weiss, J.P.; Gioannini, T.L. Molecular basis of reduced potency of underacylated endotoxins. J. Immunol. 2005, 175, 4669–4676. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Fei, N.; Zhao, L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013, 7, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Michail, S.; Lin, M.; Frey, M.R.; Fanter, R.; Paliy, O.; Hilbush, B.; Reo, N.V. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol. Ecol. 2015, 91, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gophna, U.; Sommerfeld, K.; Gophna, S.; Doolittle, W.F.; Veldhuyzen van Zanten, S.J. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 2006, 44, 4136–4141. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Lepage, P.; Nolte, A.; Hellmig, S.; Schreiber, S.; Ott, S.J. Transcriptional activity of the dominant gut mucosal microbiota in chronic inflammatory bowel disease patients. J. Med. Microbiol. 2010, 59, 1114–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartor, R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology 2008, 134, 577–594. [Google Scholar] [CrossRef] [PubMed]
- Sonnenborn, U. Escherichia coli strain Nissle 1917-from bench to bedside and back: History of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Grozdanov, L.; Zahringer, U.; Blum-Oehler, G.; Brade, L.; Henne, A.; Knirel, Y.A.; Schombel, U.; Schulze, J.; Sonnenborn, U.; Gottschalk, G.; et al. A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J. Bacteriol. 2002, 184, 5912–5925. [Google Scholar] [CrossRef] [PubMed]
- Henker, J.; Laass, M.; Blokhin, B.M.; Bolbot, Y.K.; Maydannik, V.G.; Elze, M.; Wolff, C.; Schulze, J. The probiotic Escherichia coli strain Nissle 1917 (EcN) stops acute diarrhoea in infants and toddlers. Eur. J. Pediatr. 2007, 166, 311–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandasamy, S.; Vlasova, A.N.; Fischer, D.; Kumar, A.; Chattha, K.S.; Rauf, A.; Shao, L.; Langel, S.N.; Rajashekara, G.; Saif, L.J. Differential Effects of Escherichia coli Nissle and Lactobacillus rhamnosus Strain GG on Human Rotavirus Binding, Infection, and B Cell Immunity. J. Immunol. 2016, 196, 1780–1789. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, S.; Machnowska, P.; Assmus, J.; Sieber, M.; Pieper, R.; Schmidt, M.F.; Brockmann, G.A.; Scharek-Tedin, L.; Johne, R. Feeding of the probiotic bacterium Enterococcus faecium NCIMB 10415 differentially affects shedding of enteric viruses in pigs. Vet. Res. 2012, 43, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodinova-Zadnikova, R.; Sonnenborn, U. Effect of preventive administration of a nonpathogenic Escherichia coli strain on the colonization of the intestine with microbial pathogens in newborn infants. Biol. Neonate 1997, 71, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.; Duncker, S.; Barth, S.; Bauerfeind, R.; Gruber, A.D.; Deppenmeier, S.; Breves, G. Preventive effects of the probiotic Escherichia coli strain Nissle 1917 on acute secretory diarrhea in a pig model of intestinal infection. Dig. Dis. Sci. 2006, 51, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Splichalova, A.; Trebichavsky, I.; Rada, V.; Vlkova, E.; Sonnenborn, U.; Splichal, I. Interference of Bifidobacterium choerinum or Escherichia coli Nissle 1917 with Salmonella Typhimurium in gnotobiotic piglets correlates with cytokine patterns in blood and intestine. Clin. Exp. Immunol. 2011, 163, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Kruis, W.; Fric, P.; Pokrotnieks, J.; Lukas, M.; Fixa, B.; Kascak, M.; Kamm, M.A.; Weismueller, J.; Beglinger, C.; Stolte, M.; et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 2004, 53, 1617–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthes, H.; Krummenerl, T.; Giensch, M.; Wolff, C.; Schulze, J. Clinical trial: Probiotic treatment of acute distal ulcerative colitis with rectally administered Escherichia coli Nissle 1917 (EcN). BMC Complement. Altern. Med. 2010, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Rembacken, B.J.; Snelling, A.M.; Hawkey, P.M.; Chalmers, D.M.; Axon, A.T. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: A randomised trial. Lancet 1999, 354, 635–639. [Google Scholar] [CrossRef]
- Scaldaferri, F.; Gerardi, V.; Mangiola, F.; Lopetuso, L.R.; Pizzoferrato, M.; Petito, V.; Papa, A.; Stojanovic, J.; Poscia, A.; Cammarota, G.; et al. Role and mechanisms of action of Escherichia coli Nissle 1917 in the maintenance of remission in ulcerative colitis patients: An update. World J. Gastroenterol. 2016, 22, 5505–5511. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Friesen, L.; Park, J.; Kim, H.M.; Kim, C.H. Microbial metabolites, short-chain fatty acids, restrain tissue bacterial load, chronic inflammation, and associated cancer in the colon of mice. Eur. J. Immunol. 2018, 48, 1235–1247. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llewellyn, A.; Foey, A. Probiotic Modulation of Innate Cell Pathogen Sensing and Signaling Events. Nutrients 2017, 9, 1156. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V.; Evans, C.T.; Goldstein, E.J.C. Strain-Specificity and Disease-Specificity of Probiotic Efficacy: A Systematic Review and Meta-Analysis. Front. Med. 2018, 5, 124. [Google Scholar] [CrossRef] [PubMed]
- Worthley, D.L.; Le Leu, R.K.; Whitehall, V.L.; Conlon, M.; Christophersen, C.; Belobrajdic, D.; Mallitt, K.A.; Hu, Y.; Irahara, N.; Ogino, S.; et al. A human, double-blind, placebo-controlled, crossover trial of prebiotic, probiotic, and synbiotic supplementation: Effects on luminal, inflammatory, epigenetic, and epithelial biomarkers of colorectal cancer. Am. J. Clin. Nutr. 2009, 90, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Cushing, K.; Alvarado, D.M.; Ciorba, M.A. Butyrate and Mucosal Inflammation: New Scientific Evidence Supports Clinical Observation. Clin. Transl. Gastroenterol. 2015, 6, e108. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef] [PubMed]
- Biavati, B.; Mattarelli, P. Bifidobacterium. In Bergley’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Rainey, F., Kämpfer, P., Trujillo, M., Chun, J., DeVos, P., Hedlund, B., Dedysh, S., Eds.; Wiley: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Bottacini, F.; van Sinderen, D.; Ventura, M. Omics of bifidobacteria: Research and insights into their health-promoting activities. Biochem. J. 2017, 474, 4137–4152. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; Mora, D.; Eijlander, R.T.; Wels, M.; de Vos, W.M. Comparative genomic analysis of the multispecies probiotic-marketed product VSL#3. PLoS ONE 2018, 13, e0192452. [Google Scholar]
- Douillard, F.P.; Ribbera, A.; Kant, R.; Pietila, T.E.; Jarvinen, H.M.; Messing, M.; Randazzo, C.L.; Paulin, L.; Laine, P.; Ritari, J.; et al. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet. 2013, 9, e1003683. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Armanini, F.; Turroni, F.; James, K.; Ferretti, P.; Gorfer, V.; Ferrario, C.; Milani, C.; et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 2017, 5, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, H.; Kushiro, A.; Ishikawa, E.; Kubota, H.; Gawad, A.; Sakai, T.; Oishi, K.; Martin, R.; Ben-Amor, K.; Knol, J.; et al. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS ONE 2013, 8, e78331. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Mancabelli, L.; Lugli, G.A.; Duranti, S.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Ferretti, P.; Gorfer, V.; et al. Exploring Vertical Transmission of Bifidobacteria from Mother to Child. Appl. Environ. Microbiol. 2015, 81, 7078–7087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korpela, K.; Costea, P.; Coelho, L.P.; Kandels-Lewis, S.; Willemsen, G.; Boomsma, D.I.; Segata, N.; Bork, P. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018, 28, 561–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollister, E.B.; Riehle, K.; Luna, R.A.; Weidler, E.M.; Rubio-Gonzales, M.; Mistretta, T.A.; Raza, S.; Doddapaneni, H.V.; Metcalf, G.A.; Muzny, D.M.; et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome 2015, 3, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed]
- Aakko, J.; Kumar, H.; Rautava, S.; Wise, A.; Autran, C.; Bode, L.; Isolauri, E.; Salminen, S. Human milk oligosaccharide categories define the microbiota composition in human colostrum. Benef. Microbes 2017, 8, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Bottacini, F.; O’Connell-Motherway, M.; Ryan, C.A.; Ross, R.P.; van Sinderen, D.; Stanton, C. Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genom. 2018, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, A.; Bottacini, F.; O’Connell Motherway, M.; van Sinderen, D. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genom. 2015, 16, 832. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.A.; Chapman, J.; Adeuya, A.; Kim, J.H.; Chen, F.; Whitehead, T.R.; Lapidus, A.; Rokhsar, D.S.; Lebrilla, C.B.; German, J.B.; et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 18964–18969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milani, C.; Lugli, G.A.; Duranti, S.; Turroni, F.; Mancabelli, L.; Ferrario, C.; Mangifesta, M.; Hevia, A.; Viappiani, A.; Scholz, M.; et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 2015, 5, 15782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falony, G.; Vlachou, A.; Verbrugghe, K.; De Vuyst, L. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 2006, 72, 7835–7841. [Google Scholar] [CrossRef] [PubMed]
- Moens, F.; Weckx, S.; De Vuyst, L. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int. J. Food Microbiol. 2016, 231, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Rios-Covian, D.; Gueimonde, M.; Duncan, S.H.; Flint, H.J.; de los Reyes-Gavilan, C.G. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Serafini, F.; Foroni, E.; Duranti, S.; O’Connell Motherway, M.; Taverniti, V.; Mangifesta, M.; Milani, C.; Viappiani, A.; Roversi, T.; et al. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc. Natl. Acad. Sci. USA 2013, 110, 11151–11156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell Motherway, M.; Zomer, A.; Leahy, S.C.; Reunanen, J.; Bottacini, F.; Claesson, M.J.; O’Brien, F.; Flynn, K.; Casey, P.G.; Munoz, J.A.; et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. USA 2011, 108, 11217–11222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fanning, S.; Hall, L.J.; Cronin, M.; Zomer, A.; MacSharry, J.; Goulding, D.; Motherway, M.O.; Shanahan, F.; Nally, K.; Dougan, G.; et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. USA 2012, 109, 2108–2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.Y.; Osaka, T.; Moriyama, E.; Date, Y.; Kikuchi, J.; Tsuneda, S. Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol. Rep. 2015, 3, e12327. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Varela, L.; Alonso-Guervos, M.; Garcia-Suarez, O.; Gueimonde, M.; Ruas-Madiedo, P. Screening of Bifidobacteria and Lactobacilli Able to Antagonize the Cytotoxic Effect of Clostridium difficile upon Intestinal Epithelial HT29 Monolayer. Front. Microbiol. 2016, 7, 577. [Google Scholar] [CrossRef] [PubMed]
- Srutkova, D.; Schwarzer, M.; Hudcovic, T.; Zakostelska, Z.; Drab, V.; Spanova, A.; Rittich, B.; Kozakova, H.; Schabussova, I. Bifidobacterium longum CCM 7952 Promotes Epithelial Barrier Function and Prevents Acute DSS-Induced Colitis in Strictly Strain-Specific Manner. PLoS ONE 2015, 10, e0134050. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Mangifesta, M.; Mancabelli, L.; Lugli, G.A.; Mancino, W.; Viappiani, A.; Faccini, A.; van Sinderen, D.; Ventura, M.; Turroni, F. The Sortase-Dependent Fimbriome of the Genus Bifidobacterium: Extracellular Structures with Potential To Modulate Microbe-Host Dialogue. Appl. Environ. Microbiol. 2017, 83, e01295-17. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Reunanen, J.; Meijerink, M.; Pietila, T.E.; Kainulainen, V.; Klievink, J.; Huuskonen, L.; Aalvink, S.; Skurnik, M.; Boeren, S.; et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE 2017, 12, e0173004. [Google Scholar] [CrossRef] [PubMed]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Chelakkot, C.; Choi, Y.; Kim, D.K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.S.; Jee, Y.K.; Gho, Y.S.; et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mole. Med. 2018, 50, e450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breyner, N.M.; Michon, C.; de Sousa, C.S.; Vilas Boas, P.B.; Chain, F.; Azevedo, V.A.; Langella, P.; Chatel, J.M. Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii Shows a Protective Effect on DNBS and DSS-Induced Colitis Model in Mice through Inhibition of NF-kappaB Pathway. Front. Microbiol. 2017, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Song, K.; Zhang, Y.; Zhu, C.; Shen, Z.; Wu, S.; Luo, W.; Tan, B.; Yang, Z.; Wang, X. Roseburia intestinalis-derived flagellin is a negative regulator of intestinal inflammation. Biochem. Biophys. Res. Commun. 2018, 501, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Giardino Torchia, M.L.; Lawson, G.W.; Karp, C.L.; Ashwell, J.D.; Mazmanian, S.K. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 2012, 12, 509–520. [Google Scholar] [CrossRef] [PubMed]
- AlFaleh, K.; Anabrees, J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst. Rev. 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshpande, G.; Jape, G.; Rao, S.; Patole, S. Benefits of probiotics in preterm neonates in low-income and medium-income countries: A systematic review of randomised controlled trials. BMJ Open 2017, 7, e017638. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Marwah, G.; Westgarth, M.; Buys, N.; Ellwood, D.; Gray, P.H. Effects of Probiotics on Necrotizing Enterocolitis, Sepsis, Intraventricular Hemorrhage, Mortality, Length of Hospital Stay, and Weight Gain in Very Preterm Infants: A Meta-Analysis. Adv. Nutr. 2017, 8, 749–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Martinez, M.I.; Calabuig-Tolsa, R.; Cauli, O. The effect of probiotics as a treatment for constipation in elderly people: A systematic review. Arch. Gerontol. Geriatr. 2017, 71, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.E.; Lehtoranta, L.; Lehtinen, M.J. The Effect of Bifidobacterium animalis ssp. lactis HN019 on Cellular Immune Function in Healthy Elderly Subjects: Systematic Review and Meta-Analysis. Nutrients 2017, 9, E191. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evolut. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Geerlings, S.Y.; Aalvink, S.; de Vos, W.M.; Belzer, C. Action and function of Akkermansia muciniphila in microbiome ecology, health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Ouwerkerk, J.P.; van der Ark, K.C.; Davids, M.; Claassens, N.J.; Robert Finestra, T.; de Vos, W.M.; Belzer, C. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl. Environ. Microbiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Belzer, C.; Chia, L.W.; Aalvink, S.; Chamlagain, B.; Piironen, V.; Knol, J.; de Vos, W.M. Microbial Metabolic Networks at the Mucus Layer Lead to Diet-Independent Butyrate and Vitamin B12 Production by Intestinal Symbionts. mBio 2017, 8, e000770-17. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, C.L.; Onnerfalt, J.; Xu, J.; Molin, G.; Ahrne, S.; Thorngren-Jerneck, K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity 2012, 20, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Png, C.W.; Linden, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Rajilic-Stojanovic, M.; Shanahan, F.; Guarner, F.; de Vos, W.M. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm. Bowel Dis. 2013, 19, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Dorffel, Y.; Loening-Baucke, V.; Theissig, F.; Ruckert, J.C.; Ismail, M.; Rau, W.A.; Gaschler, D.; Weizenegger, M.; Kuhn, S.; et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 2011, 60, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE 2013, 8, e71108. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, S.; Vanhoutte, P.M.; Woo, C.W.; Xu, A. Akkermansia Muciniphila Protects Against Atherosclerosis by Preventing Metabolic Endotoxemia-Induced Inflammation in Apoe-/- Mice. Circulation 2016, 133, 2434–2446. [Google Scholar] [CrossRef] [PubMed]
- Reunanen, J.; Kainulainen, V.; Huuskonen, L.; Ottman, N.; Belzer, C.; Huhtinen, H.; de Vos, W.M.; Satokari, R. Akkermansia muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl. Environ. Microbiol. 2015, 81, 3655–3662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, C.S.; Ban, M.; Choi, E.J.; Moon, H.G.; Jeon, J.S.; Kim, D.K.; Park, S.K.; Jeon, S.G.; Roh, T.Y.; Myung, S.J.; et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS ONE 2013, 8, e76520. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Huuskonen, L.; Reunanen, J.; Boeren, S.; Klievink, J.; Smidt, H.; Belzer, C.; de Vos, W.M. Characterization of Outer Membrane Proteome of Akkermansia muciniphila Reveals Sets of Novel Proteins Exposed to the Human Intestine. Front. Microbiol. 2016, 7, 1157. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tap, J.; Mondot, S.; Levenez, F.; Pelletier, E.; Caron, C.; Furet, J.P.; Ugarte, E.; Munoz-Tamayo, R.; Paslier, D.L.; Nalin, R.; et al. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 2009, 11, 2574–2584. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Flint, H.J. Faecalibacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Vos, P., Garrity, G., Jones, D., Krieg, N.R., Ludwig, W., Rainey, F.A., Schleifer, K.-H., Whitman, W., Eds.; John Wiley & Sons: New York, NY, USA, 2015. [Google Scholar]
- Lopez-Siles, M.; Khan, T.M.; Duncan, S.H.; Harmsen, H.J.; Garcia-Gil, L.J.; Flint, H.J. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl. Environ. Microbiol. 2012, 78, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Halder, C.V.; Faria, A.V.S.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Martinez-Medina, M.; Suris-Valls, R.; Aldeguer, X.; Sabat-Mir, M.; Duncan, S.H.; Flint, H.J.; Garcia-Gil, L.J. Changes in the Abundance of Faecalibacterium prausnitzii Phylogroups I and II in the Intestinal Mucosa of Inflammatory Bowel Disease and Patients with Colorectal Cancer. Inflamm. Bowel Dis. 2016, 22, 28–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Dore, J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Hooda, S.; Boler, B.M.; Serao, M.C.; Brulc, J.M.; Staeger, M.A.; Boileau, T.W.; Dowd, S.E.; Fahey, G.C., Jr.; Swanson, K.S. 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J. Nutr. 2012, 142, 1259–1265. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of inulin on the human gut microbiota: Stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 2009, 101, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Wrzosek, L.; Miquel, S.; Noordine, M.L.; Bouet, S.; Joncquel Chevalier-Curt, M.; Robert, V.; Philippe, C.; Bridonneau, C.; Cherbuy, C.; Robbe-Masselot, C.; et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Miquel, S.; Chain, F.; Natividad, J.M.; Jury, J.; Lu, J.; Sokol, H.; Theodorou, V.; Bercik, P.; Verdu, E.F.; et al. Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 2015, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, A.H.; Yakymenko, O.; Olivier, I.; Hakansson, F.; Postma, E.; Keita, A.V.; Soderholm, J.D. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand. J. Gastroenterol. 2013, 48, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Leclerc, M.; Martin, R.; Chain, F.; Lenoir, M.; Raguideau, S.; Hudault, S.; Bridonneau, C.; Northen, T.; Bowen, B.; et al. Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. mBio 2015, 6, e00300-15. [Google Scholar] [CrossRef] [PubMed]
- Quevrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermudez-Humaran, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M.; et al. Functional Characterization of Novel Faecalibacterium prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the Use of F. prausnitzii as a Next-Generation Probiotic. Front. Microbiol. 2017, 8, 1226. [Google Scholar] [CrossRef] [PubMed]
- Bircher, L.; Geirnaert, A.; Hammes, F.; Lacroix, C.; Schwab, C. Effect of cryopreservation and lyophilization on viability and growth of strict anaerobic human gut microbes. Microb. Biotechnol. 2018, 11, 721–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olle, B. Medicines from microbiota. Nat. Biotechnol. 2013, 31, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Hold, G.L.; Barcenilla, A.; Stewart, C.S.; Flint, H.J. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evolut. Microbiol. 2002, 52, 1615–1620. [Google Scholar]
- Duncan, S.H.; Aminov, R.I.; Scott, K.P.; Louis, P.; Stanton, T.B.; Flint, H.J. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int. J. Syst. Evolut. Microbiol. 2006, 56, 2437–2441. [Google Scholar] [CrossRef] [PubMed]
- Stanton, T.B.; Savage, D.C. Colonization of gnotobiotic mice by Roseburia cecicola, a motile, obligately anaerobic bacterium from murine ceca. Appl. Environ. Microbiol. 1983, 45, 1677–1684. [Google Scholar] [PubMed]
- Hold, G.L.; Schwiertz, A.; Aminov, R.I.; Blaut, M.; Flint, H.J. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 2003, 69, 4320–4324. [Google Scholar] [CrossRef] [PubMed]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Nishida, A.; Fujimoto, T.; Fujii, M.; Shioya, M.; Imaeda, H.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Andoh, A. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn’s Disease. Digestion 2016, 93, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van de Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, T.W.; Pham, H.P.; Bridonneau, C.; Aubry, C.; Lamas, B.; Martin-Gallausiaux, C.; Moroldo, M.; Rainteau, D.; Lapaque, N.; Six, A.; et al. Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. ISME J. 2016, 10, 460–477. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Song, K.; Shen, Z.; Quan, Y.; Tan, B.; Luo, W.; Wu, S.; Tang, K.; Yang, Z.; Wang, X. Roseburia intestinalis inhibits interleukin17 excretion and promotes regulatory T cells differentiation in colitis. Mol. Med. Rep. 2018, 17, 7567–7574. [Google Scholar] [PubMed]
- Shen, Z.; Zhu, C.; Quan, Y.; Yang, J.; Yuan, W.; Yang, Z.; Wu, S.; Luo, W.; Tan, B.; Wang, X. Insights into Roseburia intestinalis which alleviates experimental colitis pathology by inducing anti-inflammatory responses. J. Gastroenterol. Hepatol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.M.; Van de Wiele, T. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Young, P.; Holtrop, G.; Flint, H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 2010, 12, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Wade, W.G. Eubacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Vos, P., Garrity, G., Jones, D., Krieg, N.R., Ludwig, W., Rainey, F.A., Schleifer, K.-H., Whitman, W., Eds.; John Wiley and Sons: New York, NY, USA, 2015. [Google Scholar]
- Hirano, A.; Umeno, J.; Okamoto, Y.; Shibata, H.; Ogura, Y.; Moriyama, T.; Torisu, T.; Fujioka, S.; Fuyuno, Y.; Kawarabayasi, Y.; et al. Comparison of the microbial community structure between inflamed and non-inflamed sites in patients with ulcerative colitis. J. Gastroenterol. Hepatol. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Fekry, M.I.; Engels, C.; Zhang, J.; Schwab, C.; Lacroix, C.; Sturla, S.J.; Chassard, C. The strict anaerobic gut microbe Eubacterium hallii transforms the carcinogenic dietary heterocyclic amine 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Environ. Microbiol. Rep. 2016, 8, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.P.; Martin, J.C.; Duncan, S.H.; Flint, H.J. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol. Ecol. 2014, 87, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef] [PubMed]
- Moens, F.; Verce, M.; De Vuyst, L. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int. J. Food Microbiol. 2017, 241, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Bunesova, V.; Lacroix, C.; Schwab, C. Mucin Cross-Feeding of Infant Bifidobacteria and Eubacterium hallii. Microb. Ecol. 2018, 75, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Engels, C.; Ruscheweyh, H.J.; Beerenwinkel, N.; Lacroix, C.; Schwab, C. The Common Gut Microbe Eubacterium hallii also Contributes to Intestinal Propionate Formation. Front. Microbiol. 2016, 7, 713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zitomersky, N.L.; Coyne, M.J.; Comstock, L.E. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect. Immun. 2011, 79, 2012–2020. [Google Scholar] [CrossRef] [PubMed]
- Wexler, A.G.; Goodman, A.L. An insider’s perspective: Bacteroides as a window into the microbiome. Nature Microbiol. 2017, 2, 17026. [Google Scholar] [CrossRef] [PubMed]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed]
- Erturk-Hasdemir, D.; Kasper, D.L. Finding a needle in a haystack: Bacteroides fragilis polysaccharide A as the archetypical symbiosis factor. Ann. N. Y. Acad. Sci. 2018, 1417, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Meng, D.; Weng, M.; Zhu, W.; Wu, W.; Kasper, D.; Walker, W.A. The symbiotic bacterial surface factor polysaccharide A on Bacteroides fragilis inhibits IL-1beta-induced inflammation in human fetal enterocytes via toll receptors 2 and 4. PLoS ONE 2017, 12, e0172738. [Google Scholar]
- Chang, Y.C.; Ching, Y.H.; Chiu, C.C.; Liu, J.Y.; Hung, S.W.; Huang, W.C.; Huang, Y.T.; Chuang, H.L. TLR2 and interleukin-10 are involved in Bacteroides fragilis-mediated prevention of DSS-induced colitis in gnotobiotic mice. PLoS ONE 2017, 12, e0180025. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Li, Z.; Tan, Y.; Guo, Z.; Liu, Y.; Wang, Y.; Yuan, Y.; Yang, R.; Bi, Y.; Bai, Y.; et al. A novel strain of Bacteroides fragilis enhances phagocytosis and polarises M1 macrophages. Sci. Rep. 2016, 6, 29401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maerz, J.K.; Steimle, A.; Lange, A.; Bender, A.; Fehrenbacher, B.; Frick, J.S. Outer membrane vesicles blebbing contributes to B. vulgatus mpk-mediated immune response silencing. Gut Microbes 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Waidmann, M.; Bechtold, O.; Frick, J.S.; Lehr, H.A.; Schubert, S.; Dobrindt, U.; Loeffler, J.; Bohn, E.; Autenrieth, I.B. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 2003, 125, 162–177. [Google Scholar] [CrossRef]
- Xu, J.; Mahowald, M.A.; Ley, R.E.; Lozupone, C.A.; Hamady, M.; Martens, E.C.; Henrissat, B.; Coutinho, P.M.; Minx, P.; Latreille, P.; et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007, 5, e156. [Google Scholar] [CrossRef] [PubMed]
- Heini, T.; Kainulainen, V.; Hiippala, K.; Satokari, R. Adhesion ja anti-inflammatory properties of selected bacteroidales isolates from healthy human microbiota. In Proceedings of the Abstract and Poster in Nutrition Winter School: Breaking Barriers: Gut, Brain, Bugs—And Beyond, Levi, Finland, 22–26 January 2018. [Google Scholar]
- Jalanka, J.; Mattila, E.; Jouhten, H.; Hartman, J.; de Vos, W.M.; Arkkila, P.; Satokari, R. Long-term effects on luminal and mucosal microbiota and commonly acquired taxa in faecal microbiota transplantation for recurrent Clostridium difficile infection. BMC Med. 2016, 14, 155. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Zhu, A.; Benes, V.; Costea, P.I.; Hercog, R.; Hildebrand, F.; Huerta-Cepas, J.; Nieuwdorp, M.; Salojarvi, J.; Voigt, A.Y.; et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 2016, 352, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; DuPont, H.L. New approaches for bacteriotherapy: Prebiotics, new-generation probiotics, and synbiotics. Clin. Infect. Dis. 2015, 60 (Suppl. 2), S108–S121. [Google Scholar] [CrossRef] [PubMed]
- Holscher, H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef] [PubMed]


| Organism | Effector Molecule | Mediated Effect | Study Type | Reference |
|---|---|---|---|---|
| B. breve UCC2003 | IVb tight adherence (Tad) pili | Host colonization and persistence mechanism | in silico, in vivo | [76] |
| B. breve UCC2003 | Surface exopolysaccharide (EPS) | Acid and bile tolerance, immunomodulation, protection against pathogen colonization and burden | in silico, in vitro, in vivo | [77] |
| Several bifidobacterial strains | Sortase-dependent pili | Adhesion to host mucus components | in silico, in vitro, in vivo | [82] |
| B. bifidum PRL2010 | Sortase-dependent pili | Autoaggregation, evoked TNF-α expression and a significantly lower IL-10 response | in vitro, in vivo | [75] |
| B. longum ssp. longum CCM 7952 | Ligands for NOD2 | Reduced clinical symptoms in a colitis mouse model, preserved expression of tight junction proteins | in vitro, in vivo | [81] |
| A. muciniphila MucT | Amuc_1100 (OM pili-like protein) | Induction of cytokine production, interactionwith TLR 2, improves gut barrier | in vitro | [83,84] |
| A. muciniphila ATCC BA-835 | Extracellular vesicles | Improves intestinal barrier integrity, improved body weight & glucose tolerance | in vitro, in vivo | [85] |
| F. prausnitzii A2-165 | Microbial anti-inflammatory molecule (MAM) | Peptides of MAM inhibit the activation of NF-κB, ameliorate colitis symptoms in mice | in vitro, in vivo | [86] |
| R. intestinalis DSMZ-14610 | Flagellin | Upregulation of IncRNA, alleviation of colonic inflammation | in vitro, in vivo | [87] |
| B. fragilis NCTC 9343 | Polysaccharide A | Immune activation, elicits cytokine production, protects and treats colitis in an animal model | in vitro, in vivo | [88] |
| B. fragilis 323-J-86 | Outer-membrane vesicle | Transports PSA molecule to immune cells | in vitro, in vivo | [89] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients 2018, 10, 988. https://0-doi-org.brum.beds.ac.uk/10.3390/nu10080988
Hiippala K, Jouhten H, Ronkainen A, Hartikainen A, Kainulainen V, Jalanka J, Satokari R. The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients. 2018; 10(8):988. https://0-doi-org.brum.beds.ac.uk/10.3390/nu10080988
Chicago/Turabian StyleHiippala, Kaisa, Hanne Jouhten, Aki Ronkainen, Anna Hartikainen, Veera Kainulainen, Jonna Jalanka, and Reetta Satokari. 2018. "The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation" Nutrients 10, no. 8: 988. https://0-doi-org.brum.beds.ac.uk/10.3390/nu10080988