Extra Virgin Olive Oil Phenol Extracts Exert Hypocholesterolemic Effects through the Modulation of the LDLR Pathway: In Vitro and Cellular Mechanism of Action Elucidation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Selection of the Extra Virgin Olive Oils
2.3. HPLC-DAD Analysis of Phenols from EVOO
2.4. Preparation of the Phenolic Extracts for the Biological Testing
2.5. Analysis of the Phenolic Extract
2.6. Cell Culture Conditions and Treatment
2.7. MTT Assay
2.8. HMGCoAR Activity Assay
2.9. Western Blot Analysis
2.10. In-Cell Western (ICW) Assay
2.11. Ffluorescent LDL Uptake
2.12. Statistical Analysis
3. Results
3.1. Characterization of The Phenolic Extracts
3.2. The Phenolic Extracts Drop In Vitro the HMGCoAR Activity
3.3. Effects of the Phenols Extracts on the HepG2 Cell Vitality
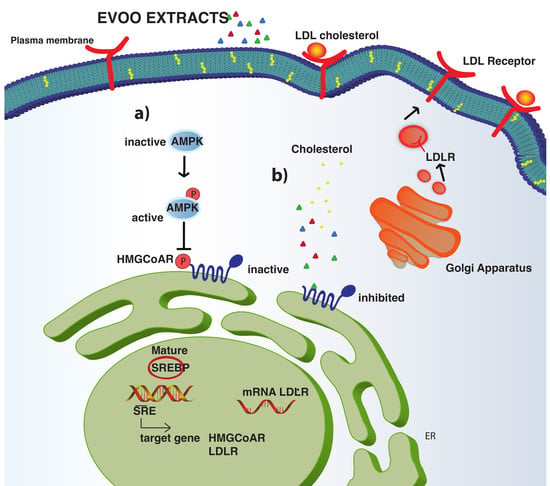
3.4. The Phenol Extracts Modulate the LDLR Pathways
3.5. The Phenol Extracts Modulate the HMGCoAR Activation by AMPK-Pathway Regulation
3.6. The Phenol Extracts Ameliorate HepG2 Ability to Absorb LDL
3.7. The Phenol Extracts Do Not Modulate the Mature PCSK9 Protein Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPK | 5’ AMP-activated protein kinase |
| BSA | Bovine serum albumin |
| CVD | Cardiovascular disease |
| DMEM | Dulbecco’s modified Eagle’s medium |
| EVOO | extra virgin olive oil |
| FBS | Fetal bovine serum |
| HepG2 | Human hepatoma G2 |
| HMGCoAR | 3-hydroxy-3-methylglutaryl Co-Enzyme A Reductase |
| HNF1-α | Hepatocyte Nuclear Factor 1 alpha |
| HPLC-DAD | high-performance liquid chromatography with a diode-array detector |
| HRP | Horseradish peroxidase |
| ICW | In-cell western |
| LDL | Low density lipoprotein |
| LDL-C | LDL cholesterol |
| LDLR | LDL receptor |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NMR | nuclear magnetic resonance |
| PBS | Phosphate buffered saline |
| PCSK9 | Proprotein Convertase Subtilisin/Kexin 9 |
| PMSF | Phenylmethanesulfonyl fluoride |
| RP | reverse phase |
| RT | Room temperature |
| SD | standard deviation |
| SDS | Sodium Dodecyl Sulphate |
| SREBP | Sterol Regulatory Element Binding Proteins |
References
- Brown, M.S.; Goldstein, J.L. Lipoprotein receptors in the liver. Control signals for plasma cholesterol traffic. J. Clin. Investig. 1983, 72, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.M.; Zhang, D.W. Hypercholesterolemia, low density lipoprotein receptor and proprotein convertase subtilisin/kexin-type 9. J. Biomed. Res. 2015, 29, 356–361. [Google Scholar] [PubMed] [Green Version]
- Goldstein, J.L.; Brown, M.S. Regulation of the mevalonate pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef]
- Lagace, T.A.; Curtis, D.E.; Garuti, R.; McNutt, M.C.; Park, S.W.; Prather, H.B.; Horton, J.D. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Investig. T 2006, 116, 2995–3005. [Google Scholar] [CrossRef] [Green Version]
- Park, S.W.; Moon, Y.A.; Horton, J.D. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J. Biol. Chem. 2004, 279, 50630–50638. [Google Scholar] [CrossRef] [Green Version]
- Seidah, N.G.; Prat, A. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 2012, 11, 367–383. [Google Scholar] [CrossRef]
- Chaudhary, R.; Garg, J.; Shah, N.; Sumner, A. PCSK9 inhibitors: A new era of lipid lowering therapy. World J. Cardiol. 2017, 9, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Perona, J.S.; Cañizares, J.; Montero, E.; Sánchez-Domínguez, J.M.; Catalá, A.; Ruiz-Gutiérrez, V. Virgin olive oil reduces blood pressure in hypertensive elderly subjects. Clin. Nutr. 2004, 23, 1113–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Covas, M.I.; Nyyssönen, K.; Poulsen, H.E.; Kaikkonen, J.; Zunft, H.J.F.; Kiesewetter, H.; Nascetti, S. The effect of polyphenols in olive oil on heart disease risk factors: A randomized trial. Ann. Intern. Med. 2006, 145, 333–341. [Google Scholar] [CrossRef]
- Carluccio, M.A.; Massaro, M.; Scoditti, E.; De Caterina, R. Vasculoprotective potential of olive oil components. Mol. Nutr. Food Res. 2007, 51, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Berrougui, H.; Cloutier, M.; Isabelle, M.; Khalil, A. Phenolic-extract from argan oil (Argania spinosa L.) inhibits human low-density lipoprotein (LDL) oxidation and enhances cholesterol efflux from human THP-1 macrophages. Atherosclerosis 2006, 184, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Marcelino, G.; Hiane, P.A.; Freitas, K.D.C.; Santana, L.F.; Pott, A.; Donadon, J.R.; & Guimarães, R.D.C.A. Effects of Olive Oil and Its Minor Components on Cardiovascular Diseases, Inflammation, and Gut Microbiota. Nutrients 2019, 11, 1826. [Google Scholar] [CrossRef] [Green Version]
- EU Regulation 1989. Off. J. Eur. Com. 2003, L295, 57–77. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex:32003R1989 (accessed on 1 April 2019).
- Bellumori, M.; Cecchi, L.; Innocenti, M.; Clodoveo, M.L.; Corbo, F.; Mulinacci, N. The EFSA Health Claim on Olive Oil Polyphenols: Acid Hydrolysis Validation and Total Hydroxytyrosol and Tyrosol Determination in Italian Virgin Olive Oils. Molecules 2019, 24, 2179. [Google Scholar] [CrossRef] [Green Version]
- IOC/T.20/Doc No. 29, Official Method of Analysis. Determination of Biophenols in Olive oil by HPLC. International Olive Council, Madrid. 2009. Available online: http://www.internationaloliveoil.org/estaticos/view/224-testing-methods (accessed on 1 April 2019).
- Bellumori, M.; Cecchi, L.; Romani, A.; Mulinacci, N.; Innocenti, M. Recovery and stability over time of phenolic fractions by an industrial filtration system of olive mill wastewaters: A three-year study. J. Sci. Food Agric. 2018, 98, 2761–2769. [Google Scholar] [CrossRef]
- Karkoula, E.; Skantzari, A.; Melliou, E.; Magiatis, P. Direct measurement of oleocanthal and oleacein levels in olive oil by quantitative (1)H NMR. Establishment of a new index for the characterization of extra virgin olive oils. J. Agric. Food Chem. 2012, 60, 11696–11703. [Google Scholar] [CrossRef]
- Zanoni, C.; Aiello, G.; Arnoldi, A.; Lammi, C. Investigations on the hypocholesterolaemic activity of LILPKHSDAD and LTFPGSAED, two peptides from lupin beta-conglutin: Focus on LDLR and PCSK9 pathways. J. Funct. Foods 2017, 32, 1–8. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Calabresi, L.; Arnoldi, A. Lupin protein exerts cholesterol-lowering effects targeting PCSK9: From clinical evidences to elucidation of the in vitro molecular mechanism using HepG2 cells. J. Funct. Foods 2016, 23, 230–240. [Google Scholar] [CrossRef]
- Lammi, C.; Zanoni, C.; Arnoldi, A. A simple and high-throughput in-cell Western assay using HepG2 cell line for investigating the potential hypocholesterolemic effects of food components and nutraceutics. Food Chem. 2015, 169, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, C.; Aiello, G.; Arnoldi, A.; Lammi, C. Hempseed peptides exert hypocholesterolemic effects with a statin-like mechanism. J. Agric. Food Chem. 2017, 65, 8829–8838. [Google Scholar] [CrossRef] [PubMed]
- Benkhalti, F.; Prost, J.; Paz, E.; Perez-Jimenez, F.; El Boustani, E. Effects of feeding virgin olive oil or their polyphenols on lipid of rat liver. Nutr. Res. 2002, 22, 1067–1075. [Google Scholar] [CrossRef]
- Istvan, E.S.; Palnitkar, M.; Buchanan, S.K.; Deisenhofer, J. Crystal structure of the catalytic portion of human HMG-CoA reductase: Insights into regulation of activity and catalysis. EMBO J. 2000, 19, 819–830. [Google Scholar] [CrossRef]
- Sun, W.; Lee, T.S.; Zhu, M.; Gu, C.; Wang, Y.; Zhu, Y.; Shyy, J.Y. Statins activate AMP-activated protein kinase in vitro and in vivo. Circulation 2006, 114, 2655–2662. [Google Scholar] [CrossRef] [Green Version]
- Levy, E.; Ouadda, A.B.D.; Spahis, S.; Sane, A.T.; Garofalo, C.; Grenier, É.; Ménard, D. PCSK9 plays a significant role in cholesterol homeostasis and lipid transport in intestinal epithelial cells. Atherosclerosis 2013, 227, 297–306. [Google Scholar] [CrossRef]
- Careskey, H.E.; Davis, R.A.; Alborn, W.E.; Troutt, J.S.; Cao, G.; Konrad, R.J. Atorvastatin increases human serum levels of proprotein convertase subtilisin/kexin type 9. J. Lipid Res. 2008, 49, 394–398. [Google Scholar] [CrossRef] [Green Version]
- Welder, G.; Zineh, I.; Pacanowski, M.A.; Troutt, J.S.; Cao, G.; Konrad, R.J. High-dose atorvastatin causes a rapid sustained increase in human serum PCSK9 and disrupts its correlation with LDL cholesterol. J. Lipid Res. 2010, 51, 2714–2721. [Google Scholar] [CrossRef] [Green Version]
- Tsimidou, M.Z.; Sotiroglou, M.; Mastralexi, A.; Nenadis, N.; García-González, D.L.; Gallina Toschi, T. In House Validated UHPLC Protocol for the Determination of the Total Hydroxytyrosol and Tyrosol Content in Virgin Olive Oil Fit for the Purpose of the Health Claim Introduced by the EC Regulation 432/2012 for “Olive Oil Polyphenols”. Molecules 2019, 24, 1044. [Google Scholar] [CrossRef] [Green Version]







| Phenolic Content | ||||
|---|---|---|---|---|
| BUO (before Hydrolysis) | OMN (before Hydrolysis) | |||
| EVOO (µg/g) | dry extract (µg/mg) | EVOO (µg/g) | dry extract (µg/mg) | |
| Free hydroxytyrosol | 9.3 ± 0.9 | 4.0 ± 0.1 | 17.4 ± 5.1 | 16.0 ± 4.6 |
| Free tyrosol | 5.1 ± 0.1 | 2.0 ± 0.1 | 15.6 ± 5.4 | 14.0 ± 0.8 |
| Total Phenols | 617.9 ± 34.1 | 289.3 ± 15.6 | 415.7 ± 7.3 | 371.1 ± 7.5 |
| BUO (after Hydrolysis) | OMN (after Hydrolysis) | |||
| EVOO (µg/g) | dry extract (µg/mg) | EVOO (µg/g) | dry extract (µg/mg) | |
| Total hydroxytyrosol | 444.9 ± 33.4 | 208.0 ± 15.6 | 169.6 ± 2.6 | 151.0 ± 3.0 |
| Total tyrosol | 332.9 ± 7.9 | 156.0 ± 3.9 | 308.6 ± 2.7 | 275.1 ± 2.7 |
| Tyr + OH-tyr | 777.8 ± 41.3 | 364.1 ± 19.5 | 478.1 ± 5.3 | 426.1 ± 5.7 |
| Integral Ratio (1H-NMR) | ||
|---|---|---|
| BUO | OMN | |
| Oleacein/Oleocanthal | 1.09 | 0.49 |
| Oleur A./Ligstr. A | 1.73 | 0.60 |
| Oleacein + Oleur. A/Oleochantal + Ligstr. A | 1.4 | 0.58 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lammi, C.; Bellumori, M.; Cecchi, L.; Bartolomei, M.; Bollati, C.; Clodoveo, M.L.; Corbo, F.; Arnoldi, A.; Mulinacci, N. Extra Virgin Olive Oil Phenol Extracts Exert Hypocholesterolemic Effects through the Modulation of the LDLR Pathway: In Vitro and Cellular Mechanism of Action Elucidation. Nutrients 2020, 12, 1723. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12061723
Lammi C, Bellumori M, Cecchi L, Bartolomei M, Bollati C, Clodoveo ML, Corbo F, Arnoldi A, Mulinacci N. Extra Virgin Olive Oil Phenol Extracts Exert Hypocholesterolemic Effects through the Modulation of the LDLR Pathway: In Vitro and Cellular Mechanism of Action Elucidation. Nutrients. 2020; 12(6):1723. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12061723
Chicago/Turabian StyleLammi, Carmen, Maria Bellumori, Lorenzo Cecchi, Martina Bartolomei, Carlotta Bollati, Maria Lisa Clodoveo, Filomena Corbo, Anna Arnoldi, and Nadia Mulinacci. 2020. "Extra Virgin Olive Oil Phenol Extracts Exert Hypocholesterolemic Effects through the Modulation of the LDLR Pathway: In Vitro and Cellular Mechanism of Action Elucidation" Nutrients 12, no. 6: 1723. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12061723