A Positive Causal Effect of Shrimp Allergy on Major Depressive Disorder Mediated by Allergy- and Immune-Related Pathways in the East Asian Population
Abstract
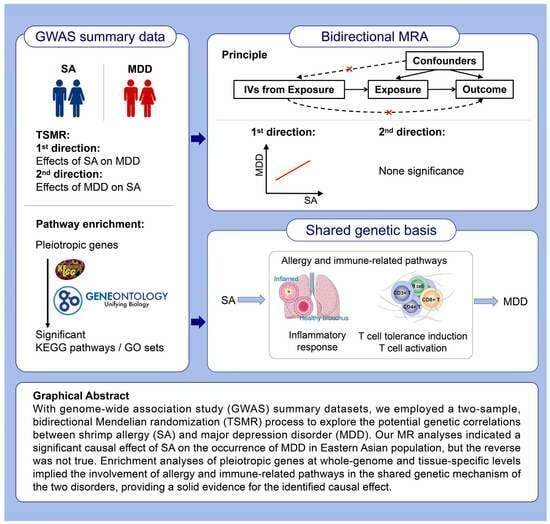
:1. Introduction
2. Materials and Methods
2.1. GWAS Summary Statistics
2.2. Two-Sample MR Study Design and Analysis
2.3. Identification of a Shared Genetic Basis between SA and MDD
2.4. Enrichment Analysis of Pleiotropic Genes for SA and MDD
3. Results
3.1. Selection of Independent Genetic Instruments
3.2. Genetic Causal Correlations between SA and MDD
3.3. Identification of a Shared Genetic Basis between SA and MDD
3.4. Identification of Pleiotropic Genes and Pathways for SA and MDD
4. Discussion
4.1. Mediation Effects of Pleiotropic Genes and Pathways on the Causal Correlation between SA and MDD
4.2. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MDD | Major Depressive Disorder |
| MR | Mendelian Randomization |
| GWAS | Genome-wide Association Study |
| SA | Shrimp Allergy |
| FA | Food Allergy |
| IgE | Immunoglobulin E |
| SNPs | Single Nucleotide Polymorphisms |
| RCT | Randomized Controlled Trial |
| MAF | Minor Allele Frequency |
| CONVERGE | China, Oxford, and Virginia Commonwealth University Experimental Research on Genetic Epidemiology |
| CKB | China Kadoorie Biobank |
| OR | Odds Ratio |
| eQTL | expression Quantitative Trait Loci |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| TSPO | Translocator Protein |
References
- Sampath, V.; Abrams, E.M.; Adlou, B.; Akdis, C.; Akdis, M.; Brough, H.A.; Chan, S.; Chatchatee, P.; Chinthrajah, R.S.; Cocco, R.R.; et al. Food allergy across the globe. J. Allergy Clin. Immunol. 2021, 148, 1347–1364. [Google Scholar] [CrossRef] [PubMed]
- De Martinis, M.; Sirufo, M.M.; Suppa, M.; Ginaldi, L. New Perspectives in Food Allergy. Int. J. Mol. Sci. 2020, 21, 1474. [Google Scholar] [CrossRef] [PubMed]
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.G.; Kim, J.-H.; Park, J.-Y.; Hwang, Y.I.; Jang, S.H.; Jung, K.-S. Association Between Asthma and Depression: A National Cohort Study. J. Allergy Clin. Immunol. Pract. 2019, 7, 1239–1245.e1. [Google Scholar] [CrossRef]
- Greenberg, P.E.; Fournier, A.-A.; Sisitsky, T.; Simes, M.; Berman, R.; Koenigsberg, S.H.; Kessler, R.C. The Economic Burden of Adults with Major Depressive Disorder in the United States (2010 and 2018). PharmacoEconomics 2021, 39, 653–665. [Google Scholar] [CrossRef]
- Proudman, D.; Greenberg, P.; Nellesen, D. The Growing Burden of Major Depressive Disorders (MDD): Implications for Researchers and Policy Makers. PharmacoEconomics 2021, 39, 619–625. [Google Scholar] [CrossRef]
- World Health Organization. Risk Assessment of Food Allergens; Part 1—Review and validation of Codex Alimentarius priority allergen list through risk assessment; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Smeekens, J.M.; Bagley, K.; Kulis, M. Tree nut allergies: Allergen homology, cross-reactivity, and implications for therapy. Clin. Exp. Allergy 2018, 48, 762–772. [Google Scholar] [CrossRef]
- Dunlop, J.H.; Keet, C.A. Epidemiology of Food Allergy. Immunol. Allergy Clin. N. Am. 2018, 38, 13–25. [Google Scholar] [CrossRef]
- Wai, C.Y.Y.; Leung, N.Y.H.; Leung, A.S.Y.; Wong, G.W.K.; Leung, T.F. Seafood Allergy in Asia: Geographical Specificity and Beyond. Front. Allergy 2021, 2, 676903. [Google Scholar] [CrossRef]
- Wamboldt, M.Z.; Schmitz, S.; Mrazek, D. Genetic association between atopy and behavioral symptoms in middle childhood. J. Child Psychol. Psychiatry 1998, 39, 1007–1016. [Google Scholar] [CrossRef]
- Timonen, M.; Jokelainen, J.; Herva, A.; Zitting, P.; Meyer-Rochow, V.B.; Räsänen, P. Presence of atopy in first-degree relatives as a predictor of a female proband’s depression: Results from the Northern Finland 1966 Birth Cohort. J. Allergy Clin. Immunol. 2003, 111, 1249–1254. [Google Scholar] [CrossRef] [PubMed]
- Galvao, P.V.M.; Silva, H.; Silva, C. Temporal distribution of suicide mortality: A systematic review. J. Affect. Disord. 2018, 228, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Timonen, M.; Viilo, K.; Hakko, H.; Särkioja, T.; Meyer-Rochow, V.B.; Väisänen, E.; Räsänen, P. Is seasonality of suicides stronger in victims with hospital-treated atopic disorders? Psychiatry Res. 2004, 126, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Uenishi, T.; Sugiura, H.; Uehara, M. Changes in the seasonal dependence of atopic dermatitis in Japan. J. Dermatol. 2001, 28, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Budu-Aggrey, A.; Joyce, S.; Davies, N.M.; Paternoster, L.; Munafò, M.R.; Brown, S.J.; Evans, J.; Sallis, H.M. Investigating the causal relationship between allergic disease and mental health. Clin. Exp. Allergy 2021, 51, 1449–1458. [Google Scholar] [CrossRef]
- Carter, A.R.; Sanderson, E.; Hammerton, G.; Richmond, R.C.; Davey Smith, G.; Heron, J.; Taylor, A.E.; Davies, N.M.; Howe, L.D. Mendelian randomisation for mediation analysis: Current methods and challenges for implementation. Eur. J. Epidemiol. 2021, 36, 465–478. [Google Scholar] [CrossRef]
- Davies, N.M.; Holmes, M.V.; Smith, G.D. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef]
- Kanchan, K.; Clay, S.; Irizar, H.; Bunyavanich, S.; Mathias, R.A. Current insights into the genetics of food allergy. J. Allergy Clin. Immunol. 2021, 147, 15–28. [Google Scholar] [CrossRef]
- Tsai, H.; Kumar, R.; Pongracic, J.; Liu, X.; Story, R.; Yu, Y.; Caruso, D.; Costello, J.; Schroeder, A.; Fang, Y.; et al. Familial aggregation of food allergy and sensitization to food allergens: A family-based study. Clin. Exp. Allergy 2009, 39, 101–109. [Google Scholar] [CrossRef]
- Pyrhönen, K.; Hiltunen, L.; Kaila, M.; Näyhä, S.; Läärä, E. Heredity of food allergies in an unselected child population: An epidemiological survey from Finland. Pediatr. Allergy Immunol. 2011, 22 Pt 2, e124–e132. [Google Scholar] [CrossRef]
- Sullivan, P.F.; Neale, M.C.; Kendler, K.S.; Bauer, A.E.; Maegbaek, M.L.; Liu, X.; Wray, N.R.; Miller, W.C.; Meltzer-Brody, S.; Munk-Olsen, T.; et al. Genetic epidemiology of major depression: Review and meta-analysis. Am. J. Psychiatry 2000, 157, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Kendall, K.M.; Van Assche, E.; Andlauer, T.F.M.; Choi, K.W.; Luykx, J.J.; Schulte, E.C.; Lu, Y. The genetic basis of major depression. Psychol. Med. 2021, 51, 2217–2230. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Larsson, S.C. An atlas on risk factors for type 2 diabetes: A wide-angled Mendelian randomisation study. Diabetologia 2020, 63, 2359–2371. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.J.; Borges, M.C.; Carnegie, R.; Mongan, D.; Rogers, P.J.; Lewis, S.J.; Thompson, A.D.; Zammit, S. Associations between plasma fatty acid concentrations and schizophrenia: A two-sample Mendelian randomisation study. Lancet Psychiatry 2021, 8, 1062–1070. [Google Scholar] [CrossRef]
- Mohammadi-Shemirani, P.; Chong, M.; Narula, S.; Perrot, N.; Conen, D.; Roberts, J.D.; Thériault, S.; Bossé, Y.; Lanktree, M.B.; Pigeyre, M.; et al. Elevated Lipoprotein(a) and Risk of Atrial Fibrillation: An Observational and Mendelian Randomization Study. J. Am. Coll. Cardiol. 2022, 79, 1579–1590. [Google Scholar] [CrossRef]
- Feng, Y.; Fu, M.; Guan, X.; Wang, C.; Yuan, F.; Bai, Y.; Meng, H.; Li, G.; Wei, W.; Li, H.; et al. Uric Acid Mediated the Association Between BMI and Postmenopausal Breast Cancer Incidence: A Bidirectional Mendelian Randomization Analysis and Prospective Cohort Study. Front. Endocrinol. 2021, 12, 742411. [Google Scholar] [CrossRef]
- Hu, X.; Rong, S.; Wang, Q.; Sun, T.; Bao, W.; Chen, L.; Liu, L. Association between plasma uric acid and insulin resistance in type 2 diabetes: A Mendelian randomization analysis. Diabetes Res. Clin. Pract. 2021, 171, 108542. [Google Scholar] [CrossRef]
- Ponce, C.B.; Scherer, D.; Brinster, R.; Boekstegers, F.; Marcelain, K.; Gárate-Calderón, V.; Müller, B.; de Toro, G.; Retamales, J.; Barajas, O.; et al. Gallstones, Body Mass Index, C-Reactive Protein, and Gallbladder Cancer: Mendelian Randomization Analysis of Chilean and European Genotype Data. Hepatology 2021, 73, 1783–1796. [Google Scholar] [CrossRef]
- Khor, S.-S.; Morino, R.; Nakazono, K.; Kamitsuji, S.; Akita, M.; Kawajiri, M.; Yamasaki, T.; Kami, A.; Hoshi, Y.; Tada, A.; et al. Genome-wide association study of self-reported food reactions in Japanese identifies shrimp and peach specific loci in the HLA-DR/DQ gene region. Sci. Rep. 2018, 8, 1069. [Google Scholar] [CrossRef]
- Giannakopoulou, O.; Lin, K.; Meng, X.; Su, M.H.; Kuo, P.H.; Peterson, R.E.; Awasthi, S.; Moscati, A.; Coleman, J.R.; Bass, N.; et al. The Genetic Architecture of Depression in Individuals of East Asian Ancestry: A Genome-Wide Association Study. JAMA Psychiatry 2021, 78, 1258–1269. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.K.; Loh, P.R.; Finucane, H.K.; Ripke, S.; Yang, J.; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M.; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, D.; Chung, Y.-J.; Xu, S. Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations. Hereditas 2018, 155, 19. [Google Scholar] [CrossRef] [PubMed]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Smith, G.D.; Davies, N.M.; Dudbridge, F.; Gill, D.; Glymour, M.M.; Hartwig, F.P.; Kutalik, Z.; Holmes, M.V.; Minelli, C.; et al. Guidelines for performing Mendelian randomization investigations: Update for summer 2023. Wellcome Open Res. 2019, 4, 186. [Google Scholar] [CrossRef] [PubMed]
- Staiger, D.; Stock, J.H. Instrumental Variables Regression with Weak Instruments. Econometrica 1997, 65, 557–586. [Google Scholar] [CrossRef]
- Chen, L.; Yang, H.; Li, H.; He, C.; Yang, L.; Lv, G. Insights into modifiable risk factors of cholelithiasis: A Mendelian randomization study. Hepatology 2022, 75, 785–796. [Google Scholar] [CrossRef]
- Hemani, G.; Tilling, K.; Davey Smith, G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017, 13, e1007081. [Google Scholar]
- Pickrell, J.K.; Berisa, T.; Liu, J.Z.; Ségurel, L.; Tung, J.Y.; Hinds, D.A. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016, 48, 709–717. [Google Scholar] [CrossRef]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef]
- Barbeira, A.N.; Dickinson, S.P.; Bonazzola, R.; Zheng, J.; Wheeler, H.E.; Torres, J.M.; Torstenson, E.S.; Shah, K.P.; Garcia, T.; Edwards, T.L.; et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018, 9, 1825. [Google Scholar] [CrossRef]
- Barbeira, A.N.; Pividori, M.; Zheng, J.; Wheeler, H.E.; Nicolae, D.L.; Im, H.K. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 2019, 15, e1007889. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Moonesinghe, H.R.; Kilburn, S.; Mackenzie, H.; Turner, P.J.; Venter, C.; Lee, K.; Dean, T. Prevalence of fish and shellfish allergy: A systematic review. J. Allergy Clin. Immunol. 2016, 133, 264–272.e4. [Google Scholar] [CrossRef]
- Rao, S.; Leung, C.S.T.; Lam, M.H.; Wing, Y.K.; Waye, M.M.Y.; Tsui, S.K.W. Resequencing three candidate genes discovers seven potentially deleterious variants susceptibility to major depressive disorder and suicide attempts in Chinese. Gene 2017, 603, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Shi, M.; Han, X.; Lam, M.H.B.; Chien, W.T.; Zhou, K.; Liu, G.; Wing, Y.K.; So, H.-C.; Waye, M.M.Y. Genome-wide copy number variation-, validation- and screening study implicates a new copy number polymorphism associated with suicide attempts in major depressive disorder. Gene 2020, 755, 144901. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Siu, C.O.; Shi, M.; Zhang, J.; Lam, M.H.B.; Yu, M.; Wing, Y.K.; Waye, M.M.Y. Associations of Homer Scaffolding Protein 1 gene and psychological correlates with suicide attempts in Chinese: A pilot study of multifactorial risk model. Gene 2018, 679, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Wray, N.R.; Ripke, S.; Mattheisen, M.; Trzaskowski, M.; Byrne, E.M.; Abdellaoui, A.; Adams, M.J.; Agerbo, E.; Air, T.M.; Andlauer, T.M.F.; et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018, 50, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Gupta, J.; Johansson, E.; Bernstein, J.A.; Chakraborty, R.; Hershey, G.K.K.; Rothenberg, M.E.; Mersha, T.B. Resolving the etiology of atopic disorders by using genetic analysis of racial ancestry. J. Allergy Clin. Immunol. 2016, 138, 676–699. [Google Scholar] [CrossRef]
- Koplin, J.J.; Allen, K.J.; Gurrin, L.C.; Peters, R.L.; Lowe, A.J.; Tan, H.-T.T.; Dharmage, S.C. The impact of family history of allergy on risk of food allergy: A population-based study of infants. Int. J. Environ. Res. Public Health 2013, 10, 5364–5377. [Google Scholar] [CrossRef]
- Hong, X.; Hao, K.; Ladd-Acosta, C.; Hansen, K.D.; Tsai, H.J.; Liu, X.; Xu, X.; Thornton, T.A.; Caruso, D.; Keet, C.A.; et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat. Commun. 2015, 6, 6304. [Google Scholar] [CrossRef]
- Chang, J.-C.; Kuo, H.-C.; Hsu, T.-Y.; Ou, C.-Y.; Liu, C.-A.; Chuang, H.; Liang, H.-M.; Huang, H.-W.; Yang, K.D. Different genetic associations of the IgE production among fetus, infancy and childhood. PLoS ONE 2013, 8, e70362. [Google Scholar] [CrossRef] [PubMed]
- Furuya, T.K.; Jacob, C.E.; Tomit, M.T.P.; Camacho, L.C.C.; Ramos, M.F.K.P.; Eluf-Neto, J.; Alves, V.A.F.; Zilberstein, B.; Cecconello, I.; Ribeiro, U., Jr.; et al. Association between Polymorphisms in Inflammatory Response-Related Genes and the Susceptibility, Progression and Prognosis of the Diffuse Histological Subtype of Gastric Cancer. Genes 2018, 9, 631. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Anumanthan, A.; Brown, J.A.; Greenfield, E.A.; Zhu, B.; Freeman, G.J. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat. Immunol. 2008, 9, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Armingol, E.; Officer, A.; Harismendy, O.; Lewis, N.E. Deciphering cell–cell interactions and communication from gene expression. Nat. Rev. Genet. 2021, 22, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, A.H.; Schwartzberg, P.L.; Joseph, R.E.; Berg, L.J. T-cell signaling regulated by the Tec family kinase, Itk. Cold Spring Harb. Perspect. Biol. 2010, 2, a002287. [Google Scholar] [CrossRef]
- Weeks, S.; Harris, R.; Karimi, M. Targeting ITK signaling for T cell-mediated diseases. iScience 2021, 24, 102842. [Google Scholar] [CrossRef]
- Liu, X.; Yu, X.; Xu, X.; Zhang, X.; Zhang, X. The protective effects of Poria cocos-derived polysaccharide CMP33 against IBD in mice and its molecular mechanism. Food Funct. 2018, 9, 5936–5949. [Google Scholar] [CrossRef]
- Wen, Y.-A.; Li, X.; Goretsky, T.; Weiss, H.L.; Barrett, T.A.; Gao, T. Loss of PHLPP protects against colitis by inhibiting intestinal epithelial cell apoptosis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2015, 1852, 2013–2023. [Google Scholar] [CrossRef]
- Zhuang, W.; Zhang, L.; Zheng, Y.; Liu, B.; Ma, C.; Zhao, W.; Liu, S.; Liu, F.; Gao, C. USP3 deubiquitinates and stabilizes the adapter protein ASC to regulate inflammasome activation. Cell. Mol. Immunol. 2022, 19, 1141–1152. [Google Scholar] [CrossRef]
- Alamuru, N.P.; Behera, S.; Butchar, J.P.; Tridandapani, S.; Suraj, S.K.; Babu, P.P.; Hasnain, S.E.; Ehtesham, N.Z.; Parsa, K.V.L. A novel immunomodulatory function of PHLPP1: Inhibition of iNOS via attenuation of STAT1 ser727 phosphorylation in mouse macrophages. J. Leukoc. Biol. 2014, 95, 775–783. [Google Scholar] [CrossRef]
- Zayed, M.A.; Jin, X.; Yang, C.; Belaygorod, L.; Engel, C.; Desai, K.; Harroun, N.; Saffaf, O.; Patterson, B.W.; Hsu, F.F.; et al. CEPT1-Mediated Phospholipogenesis Regulates Endothelial Cell Function and Ischemia-Induced Angiogenesis Through PPARalpha. Diabetes 2021, 70, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Freeland, D.M.H.; Nadeau, K.C. Food allergy: Immune mechanisms, diagnosis and immunotherapy. Nat. Rev. Immunol. 2016, 16, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Black, C.; Miller, B.J. Meta-Analysis of Cytokines and Chemokines in Suicidality: Distinguishing Suicidal Versus Nonsuicidal Patients. Biol. Psychiatry 2015, 78, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Holmes, S.E.; Hinz, R.; Conen, S.; Gregory, C.J.; Matthews, J.C.; Anton-Rodriguez, J.M.; Gerhard, A.; Talbot, P.S. Elevated Translocator Protein in Anterior Cingulate in Major Depression and a Role for Inflammation in Suicidal Thinking: A Positron Emission Tomography Study. Biol. Psychiatry 2018, 83, 61–69. [Google Scholar] [CrossRef]
- Tao, R.; Fu, Z.; Xiao, L. Chronic Food Antigen-specific IgG-mediated Hypersensitivity Reaction as A Risk Factor for Adolescent Depressive Disorder. Genom. Proteom. Bioinform. 2019, 17, 183–189. [Google Scholar] [CrossRef]






| Characters/Phenotype | Shrimp Allergy | Major Depressive Disorder |
|---|---|---|
| Abbreviation | SA | MDD |
| Cases | 539 | 15,771 |
| Controls | 8350 | 178,777 |
| Original no. of SNPs | 581,673 | 7,440,922 |
| No. of processed SNPs | 570,779 | 5,437,803 |
| Overlapped SNPs | 494,741 | |
| Ethnicity | East Asian | |
| Exposure | Outcome | No. of Clumped SNPs a | No. of SNPs in MRA b | MR p-Value c | MR Method d | Heterogeneity | Pleiotropy | Directionality |
|---|---|---|---|---|---|---|---|---|
| Independent genetic SNPs with p < 5 × 10−6 (Group 1) | ||||||||
| SA | MDD | 15 | 6 | 5.01 × 10−3 | fixed-effects IVW | No | No | TRUE |
| MDD | SA | 7 | 0 | ND e | -- | -- | -- | -- |
| Independent genetic SNPs with F > 10 (Group 2) | ||||||||
| SA | MDD | 569 | 473 | 1.21 × 10−2 | MR Egger | No | Yes | TRUE |
| MDD | SA | 1535 | 265 | 3.98 × 10−1 | fixed-effects IVW | No | No | FALSE |
| Independent genetic SNPs with p < 5 × 10−3 (Group 3) | ||||||||
| SA | MDD | 1518 | 1302 | 2.81 × 10−2 | MR Egger | No | Yes | TRUE |
| MDD | SA | 4024 | 690 | 6.87 × 10−1 | fixed-effects IVW | No | No | FALSE |
| SNP ID | Chromosome | Physical Position | PPA3 | Mapped Gene |
|---|---|---|---|---|
| rs76337386 | chr1 | 21493585 | 0.8174 | EIF4G3 |
| rs17130426 | chr1 | 88790511 | 0.8195 | - |
| rs1281337 | chr1 | 182111988 | 0.8913 | - |
| rs118061427 | chr2 | 92108070 | 1 | SLC9B1P2 |
| rs4236131 | chr6 | 55319676 | 0.8074 | HMGCLL1 |
| rs341058 | chr11 | 72385725 | 0.8581 | PDE2A |
| rs16954906 | chr13 | 98747021 | 0.8328 | - |
| Methods | GWAS Dataset | N | p < 0.05 | Overlapped Genes |
|---|---|---|---|---|
| MAGMA | SA (Whole-genome) | 16,644 | 833 | 44 (ABHD2, AFF3, C2, CCDC83, CCDC88B, CFB, CHPT1, DARS1, ECE1, FAM181B, FKBP1A, GFI1B, GNPDA2, GPR18, GPR20, HSPA13, HSPA1A, HSPA1L, ID4, IFT122, KIF3C, LSM2, LYPD6, MAP2K3, MBD4, NFIX, PEAK1, PGM2, PPP4R4, PRDM14, PTCH1, PTP4A3, R3HDML, RASAL1, ROPN1L, SEMA3D, SERPINA10, SH3TC1, SKIC2, TES, THBS2, TLE3, TMEM25, TTC36) |
| MDD (Whole-genome) | 18,162 | 1088 | ||
| MetaXcan | SA (brain) | 12,197 | 580 | 17 (C2orf42, EXOC2, FANCI, HLA-DMA, IQCG, ITK, ITLN1, LCE1C, LINC00581, MYH7B, PPP1R14B, RAMP3, RNF175, RSU1, TNFRSF14, TPSD1, VTN) |
| MDD (brain) | 12,258 | 303 | ||
| SA (whole blood) | 6383 | 305 | 6 (CEPT1, GNB1L, PHLPP1, USP3, ZNF440, ZNF670) | |
| MDD (whole blood) | 7206 | 94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, S.; Chen, X.; Ou, O.Y.; Chair, S.Y.; Chien, W.T.; Liu, G.; Waye, M.M.Y. A Positive Causal Effect of Shrimp Allergy on Major Depressive Disorder Mediated by Allergy- and Immune-Related Pathways in the East Asian Population. Nutrients 2024, 16, 79. https://0-doi-org.brum.beds.ac.uk/10.3390/nu16010079
Rao S, Chen X, Ou OY, Chair SY, Chien WT, Liu G, Waye MMY. A Positive Causal Effect of Shrimp Allergy on Major Depressive Disorder Mediated by Allergy- and Immune-Related Pathways in the East Asian Population. Nutrients. 2024; 16(1):79. https://0-doi-org.brum.beds.ac.uk/10.3390/nu16010079
Chicago/Turabian StyleRao, Shitao, Xiaotong Chen, Olivia Yanlai Ou, Sek Ying Chair, Wai Tong Chien, Guangming Liu, and Mary Miu Yee Waye. 2024. "A Positive Causal Effect of Shrimp Allergy on Major Depressive Disorder Mediated by Allergy- and Immune-Related Pathways in the East Asian Population" Nutrients 16, no. 1: 79. https://0-doi-org.brum.beds.ac.uk/10.3390/nu16010079