Bacillus thuringiensis Vip1 Functions as a Receptor of Vip2 Toxin for Binary Insecticidal Activity against Holotrichia parallela
Abstract
:1. Introduction
2. Results
2.1. Preparation of Vip1Ad and Vip2Ag
2.2. Bioassay of Vip1Ad and Vip2Ag against H. parallela Larvae
2.3. Histopathological Effects of Vip1Ad/Vip2Ag on H. parallela Larvae
2.4. Analysis of Binding of Vip1Ad and Vip2Ag to BBMVs of H. parallela
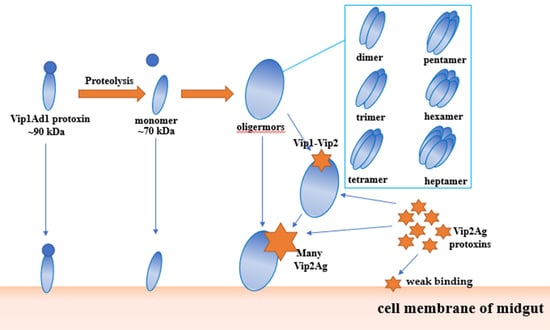
2.5. Analysis of the Interaction between Vip1Ad and Vip2Ag
2.6. Ultracentrifugation Analysis of Oligomerization of Vip1Ad-t and Vip2Ag
3. Discussion
4. Materials and Methods
4.1. Expression of Vip1Ad and Vip2Ag
4.2. Purification of Vip1Ad and Vip2Ag
4.3. Bioassay
4.4. Statistical Analysis
4.5. Preparation and Sectioning of Insect Tissues for Histopathology Observation
4.6. Preparation of BBMVs of H. parallela and Purity of BBMV Determination
4.7. Binding of Vip1Ad and Vip2Ag to BBMVs of H. parallela by Western Blotting
4.8. Protein Biotinylation
4.9. Binding of Vip1Ad and Vip2Ag to BBMVs of H. parallela by ELISA
4.10. Ligand Blot Analysis of the Interactions of Vip1Ad, Vip1Ad-t and Vip2Ag
4.11. Dot Blot Analysis of the Binding of Vip1Ad, Vip1Ad-t to Vip2Ag
4.12. Analysis of Oligomerization of Vip1Ad by Ultracentrifugation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wiley-Black, W. White grubs. EPPO Bull. 2005, 35, 229–232. [Google Scholar]
- Gibb, W.G.; Buhler, T.J. Control of white grubs at the purdue university agronomy research center. Arthropod Manag. Tests 1995, 20, 290. [Google Scholar] [CrossRef]
- Li, K.B.; Zhang, M.C.; Yin, J.; Cao, Y.Z.; Chu, J.X. Research progress on the occurrences of white grub and its control. China Plant Prot. 2014, 34, 20–28. (In Chinese) [Google Scholar]
- Bravo, A.; Likitvivatanavong, S.; Gill, S.S.; Soberón, M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 2011, 41, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, J.; Huang, D.F.; Gao, J.G.; Song, F.P. Characterization of Bacillus thuringiensis strain Bt185 toxic to the Asian cockchafer: Holotrichia parallela. Curr. Microbiol. 2006, 53, 13–17. [Google Scholar] [CrossRef]
- Shu, C.L.; Yan, G.X.; Wang, R.Y.; Zhang, J.; Feng, S.L.; Huang, D.F.; Song, F.P. Characterization of a novel cry8 gene specific to Melolonthidae pests: Holotrichia oblita and Holotrichia parallela. Appl. Microbiol. Biotechnol. 2009, 84, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zhang, Y.R.; Shu, C.L.; Crickmore, N.; Wang, Q.L.; Du, L.X.; Song, F.P.; Zhang, J. Genomic sequencing identifies novel Bacillus thuringiensis Vip1/Vip2 binary and Cry8 toxins that have high toxicity to Scarabaeoidea larvae. Appl. Microbiol. Biotechnol. 2015, 99, 753–760. [Google Scholar] [CrossRef]
- Crickmore, N.; Baum, J.; Bravo, A.; Lereclus, D.; Narva, K.; Sampson, K.; Schnepf, E.; Sun, M.; Zeigler, D.R. Bacillus thuringiensis toxin nomenclature. Bt Toxin Nomencl. 2018. Available online: http://www.btnomenclature.info/ (accessed on 1 July 2019).
- Shu, C.L.; Yu, H.; Wang, R.Y.; Feng, S.L.; Su, X.D.; Huang, D.F.; Zhang, J.; Song, F.P. Characterization of two novel cry8 genes from Bacillus thuringiensis strain Bt185. Curr. Microbiol. 2009, 4, 389–392. [Google Scholar] [CrossRef]
- Madshus, I.H.; Stenmark, H. Entry of ADP-ribosylating toxins into cells. Curr. Top. Microbiol. Immunol. 1992, 175, 1–26. [Google Scholar]
- Takehara, M.; Takagishi, T.; Seike, S.; Oda, M.; Sakaguchi, Y.; Hisatsune, J.; Ochi, S.; Kobayashi, K.; Nagahama, M. Cellular entry of Clostridium perfringens iota-toxin and Clostridium botulinum C2 toxin. Toxins 2017, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Holger, B.; Klaus, A.; Michel, R.P.; Bradley, G.S. Binary bacterial toxins: Biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol. Mol. Biol. Rev. 2004, 68, 373–402. [Google Scholar]
- Schleberger, C.; Hochmann, H.; Barth, H.; Aktories, K.; Schulz, G.E. Structure and action of the binary C2 toxin from Clostridium botulinum. J. Mol. Biol. 2006, 364, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, H.; Nagahama, M.; Nishimura, H.; Hisatsune, J.; Sakaguchi, Y.; Itogawa, Y.; Katunuma, N.; Sakurai, J. Crystal structure and site-directed mutagenesis of enzymatic components from Clostridium perfringens iota-toxin. J. Mol. Biol. 2003, 325, 471–483. [Google Scholar] [CrossRef]
- Leuber, M.; Orlik, F.; Schiffler, B.; Sickmann, A.; Benz, R. Vegetative insecticidal protein (Vip1Ac) of Bacillus thuringiensis HD201: Evidence for oligomer and channel formation. Biochemistry 2006, 45, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Jucovic, M.; Walters, F.S.; Warren, G.W.; Palekar, N.V.; Chen, J.S. From enzyme to zymogen: Engineering Vip2, an ADP-ribosyltransferase from Bacillus cereus, for conditional toxicity. Protein Eng. Des. Sel. 2008, 21, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Chakroun, M.; Banyuls, N.; Bel, Y.; Escriche, B.; Ferré, J. Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 329–350. [Google Scholar] [CrossRef]
- Lorence, A.; Darszon, A.; Bravo, A. Aminopeptidase dependent pore formation of Bacillus thuringiensis Cry1Ac toxin on Trichoplusia ni membranes. FEBS Lett. 1997, 414, 303–307. [Google Scholar] [PubMed]
- Huang, S.Z.; Shu, C.L.; Liu, C.Q.; Ding, X.Z.; Zhang, J. Screening of insecticidal activity protein against Holotrichia parallela adults from Bacillus thuringiensis. Chin. J. Biol. Control 2018, 34, 546–552. (In Chinese) [Google Scholar]
- Sattar, S.; Maiti, M.K. Molecular characterization of a novel vegetative insecticidal protein from Bacillus thuringiensis effective against sap-sucking insect pest. J. Microbiol. Biotechnol. 2011, 21, 937–946. [Google Scholar] [CrossRef]
- Stiles, B.G.; Wilkins, T.D. Purification and characterization of Clostridium perfringens iota toxin: Dependence on two nonlinked proteins for biological activity. Infect. Immun. 1986, 54, 683–688. [Google Scholar] [PubMed]
- Ohishi, I.; Iwasaki, M.; Sakaguchi, G. Purification and characterization of two components of botulinum C2 toxin. Infect. Immun. 1980, 30, 668–673. [Google Scholar] [PubMed]
- Stiles, B.G.; Barth, H.; Popoff, M.R. Clostridium perfringens iota toxin: A successfully shared template for common enteric pathogens. In Microbial Toxins; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1–20. [Google Scholar]
- Abd El-Ghany, N.M.; Saker, M.; Salama, H.S.; Ragaie, M. Histopathology of the larval midgut of Helicoverpa armigera (Hubner) fed on Bacillus thuringiensis crystals and Bt-tomato plants. J. Genet. Eng. Biotechnol. 2015, 13, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.B.; Liu, C.X.; Lu, G.Q.; Cheng, H.M.; Shen, Z.C.; Wu, K.M. Effects of Vip3AcAa+Cry1Ac cotton on midgut tissue in Helicoverpa armigera (Lepidoptera: Noctuidae). J. Insect Sci. 2018, 18, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Rouis, S.; Chakroun, M.; Saadaoui, I.; Jaoua, S. Proteolysis, histopathological effects, and immunohistopathological localization of delta-endotoxins of Bacillus thuringiensis subsp. kurstaki in the midgut of lepidopteran olive tree pathogenic insect Prays oleae. . Mol. Biotechnol. 2007, 35, 141–148. [Google Scholar] [PubMed]
- Abdelkefi-Mesrati, L.; Boukedi, H.; Dammak-Karray, M.; Sellami-Boudawara, T.; Jaoua, S.; Tounsi, S. Study of the Bacillus thuringiensis Vip3Aa16 histopathological effects and determination of its putative binding proteins in the midgut of Spodoptera littoralis. J. Invertebr. Pathol. 2011, 106, 250–254. [Google Scholar] [CrossRef]
- Torres, A.R.; Peterson, E.A. Purification of complex protein mixtures by ion-exchange displacement chromatography using spacer displacers. J. Chromatogr. 1992, 604, 39–46. [Google Scholar] [CrossRef]
- Shneider, M.M.; Buth, S.A.; Ho, B.T.; Basler, M.; Mekalanos, J.J.; Leiman, P.G. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 2013, 500, 350–353. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liang, G.M.; Zhang, L.L.; Wei, J.Z. Pathological changes in midgut tissues of larvae of the cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae), after feeding Vip3Aa protein. Acta Entomol. Sin. 2012, 55, 869–876. (In Chinese) [Google Scholar]
- Wolfersberger, M.; Luethy, P.; Maurer, A.; Parenti, P.; Sacchi, F.V.; Giordana, B.; Hanozet, G.M. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage bueetrfly (pieris brassicae). Comp. Biochem. Physiol. 1987, 86A, 301–308. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 277, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Gómez, I.; Sánchez, J.; Muñoz-Garay, C.; Matus, V.; Gill, S.S.; Soberón, M.; Bravo, A. Bacillus thuringiensis Cry1A toxins are versatile proteins with multiple modes of action: Two distinct pre-pores are involved in toxicity. Biochem. J. 2014, 459, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, M.T.; Overgaard, M.T.; Sorensen, E.S.; Stachowiak, D.; Boldt, H.B.; Kristensen, L.; Sottrup-Jensen, L.; Oxvig, C. Complex of pregnancy-associated plasma protein-A and the proform of eosinophil major basic protein. Disulfide structure and carbohydrate attachment. J. Biol. Chem. 2003, 278, 2106–2117. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.M.; Ross, N.W.; Ulmer, J.B.; Braun, P.E. Interaction of myelin basic protein and proteolipid protein. J. Neurosci. Res. 1989, 22, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Milner, R.E.; Trayhurn, P. Rapid quantitation of uncoupling protein in brown adipose tissue mitochondria by a dot immunobinding (“dot blot”) procedure: Application to the measurement of uncoupling protein in Richardson’s ground squirrel, rats, and mice. Biochem. Cell Biol. 1990, 68, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A.; Ruvinov, S.; Hess, S.; Schantz, R.; Delmer, D.P.; Wlodawer, A. Plant annexins form calcium-independent oligomers in solution. Protein Sci. 2002, 11, 2033–2040. [Google Scholar] [CrossRef] [Green Version]





| Protoxin | LC50 (μg/g soil) a | Slope ± SEM b |
|---|---|---|
| Vip1Ad | >50 | |
| Vip2Ag | >50 | |
| Vip1Ad + Vip2Ag | 2.33 (1.14–4.08) | 1.08 ± 0.21 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, J.; Jiang, J.; Shu, C.; Wang, Z.; Song, F.; Geng, L.; Duan, J.; Zhang, J. Bacillus thuringiensis Vip1 Functions as a Receptor of Vip2 Toxin for Binary Insecticidal Activity against Holotrichia parallela. Toxins 2019, 11, 440. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11080440
Geng J, Jiang J, Shu C, Wang Z, Song F, Geng L, Duan J, Zhang J. Bacillus thuringiensis Vip1 Functions as a Receptor of Vip2 Toxin for Binary Insecticidal Activity against Holotrichia parallela. Toxins. 2019; 11(8):440. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11080440
Chicago/Turabian StyleGeng, Jianxun, Jian Jiang, Changlong Shu, Zeyu Wang, Fuping Song, Lili Geng, Jiangyan Duan, and Jie Zhang. 2019. "Bacillus thuringiensis Vip1 Functions as a Receptor of Vip2 Toxin for Binary Insecticidal Activity against Holotrichia parallela" Toxins 11, no. 8: 440. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins11080440