Mechanistic Insight into the Peptide Binding Modes to Two M. tb MazF Toxins
Abstract
:1. Introduction
2. Results
2.1. The Heterologous Interactions between the MazEF Families
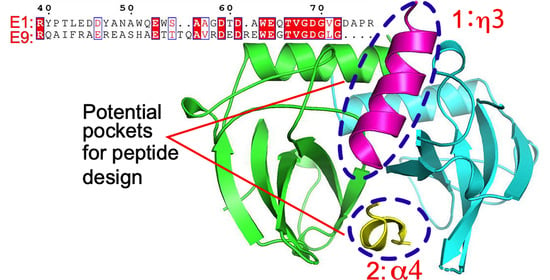
2.2. The Interactions of MazE-mt9/α4 with MazF-mt9
2.3. The Interactions of the MazE-mt1/α3 Helix with MazF-mt1
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression, and Purification of the Proteins
4.2. Crystallization, Data Collection, and Structure Determination
4.3. tRNA Cleavage Assays
4.4. Isothermal Titration Calorimetry (ITC)
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2018; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Tiwari, P.; Arora, G.; Singh, M.; Kidwai, S.; Narayan, O.; Singh, R. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 2015, 6, 6059. [Google Scholar] [CrossRef] [Green Version]
- Albrethsen, J.; Agner, J.; Piersma, S.; Højrup, P.; Pham, T.; Weldingh, K.; Jimenez, C.; Andersen, P.; Rosenkrands, I. Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol. Cell Proteom. 2013, 12, 1180–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betts, J.; Lukey, P.; Robb, L.; McAdam, R.; Duncan, K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002, 43, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Rand, L.; Hinds, J.; Springer, B.; Sander, P.; Buxton, R.; Davis, E. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol. Microbiol. 2003, 50, 1031–1042. [Google Scholar] [CrossRef]
- Denkin, S.; Byrne, S.; Jie, C.; Zhang, Y. Gene expression profiling analysis of Mycobacterium tuberculosis genes in response to salicylate. Arch. Microbiol. 2005, 184, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Provvedi, R.; Boldrin, F.; Falciani, F.; Palù, G.; Manganelli, R. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 2009, 155, 1093–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Barry, C.; Boshoff, H. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J. Bacteriol. 2010, 192, 1279–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, P.; Couturier, M.J.J.o.M.B. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Bacteriol. 1992, 226, 735–745. [Google Scholar] [CrossRef]
- Unterholzner, S.; Poppenberger, B.; Rozhon, W. Toxin-antitoxin systems: Biology, identification, and application. Mob. Genet. Elements. 2013, 3, e26219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, J.; Hergenrother, P. Artificial activation of toxin-antitoxin systems as an antibacterial strategy. Trends Microbiol. 2012, 20, 291–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sala, A.; Bordes, P.; Genevaux, P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins 2014, 6, 1002–1020. [Google Scholar] [CrossRef] [Green Version]
- Slayden, R.; Dawson, C.; Cummings, J. Toxin-antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Son, W.; Lee, B. Structural overview of toxin-antitoxin systems in infectious bacteria: A target for developing antimicrobial agents. Biochim. Biophys. Acta 2013, 1834, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Coussens, N.; Daines, D. Wake me when it’s over-Bacterial toxin-antitoxin proteins and induced dormancy. Exp. Biol. Med. 2016, 241, 1332–1342. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.; Espinosa, M.; Yeo, C. Keeping the wolves at bay: Antitoxins of prokaryotic type II toxin-antitoxin systems. Front. Mol. Biosci. 2016, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Tu, J.; Liu, Z.; Meng, F.; Ma, P.; Ding, Z.; Yang, C.; Chen, L.; Deng, X.; Xie, W. Structure of the MazF-mt9 toxin, a tRNA-specific endonuclease from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2017, 486, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Tu, J.; Tan, Y.; Cai, X.; Yang, C.; Deng, X.; Su, B.; Ma, S.; Liu, X.; Ma, P.; et al. Structural and biochemical characterization of the cognate and heterologous interactions of the MazEF-mt9 TA System. ACS Infect. Dis. 2019, 5, 1306–1316. [Google Scholar] [CrossRef]
- Chen, R.; Zhou, J.; Sun, R.; Du, C.; Xie, W. Conserved conformational changes in the regulation of MazEF-mt1. ACS Infect. Dis. 2020, 6, 1783–1795. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, Y.; Teh, J.S.; Zhang, J.; Connell, N.; Rubin, H.; Inouye, M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 18638. [Google Scholar] [CrossRef] [Green Version]
- Chopra, N.; Agarwal, S.; Verma, S.; Bhatnagar, S.; Bhatnagar, R. Modeling of the structure and interactions of the B. anthracis antitoxin, MoxX: Deletion mutant studies highlight its modular structure and repressor function. J. Comput. Aided. Mol. Des. 2011, 25, 275–291. [Google Scholar] [CrossRef]
- Kang, S.; Jin, C.; Kim, D.; Park, S.; Han, S.; Lee, B. Structure-based design of peptides that trigger Streptococcus pneumoniae cell death. FEBS J. 2020. [Google Scholar] [CrossRef]
- Minor, W.; Cymborowski, M.; Otwinowski, Z.; Maksymilian, C. Crystallography, C. HKL-3000: The integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Cryst. D Biol. Cryst. 2006, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.; Cowtan, K. Biological crystallography. Features and development of Coot. Acta Cryst. D Biol. Cryst. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonine, P.; Grosse-Kunstleve, R.; Echols, N.; Headd, J.; Moriarty, N.; Mustyakimov, M.; Terwilliger, T.; Urzhumtsev, A.; Zwart, P.; Adams, P. Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst. D Biol. Cryst. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, C.; Headd, J.; Moriarty, N.; Prisant, M.; Videau, L.; Deis, L.; Verma, V.; Keedy, D.; Hintze, B.; Chen, V.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]




| PDB IDs | MazF-mt9-α4 | MazF-mt1-α3 |
|---|---|---|
| 7DU4 | 7DU5 | |
| Data collection | SSRF BL19U1 | SSRF BL19U1 |
| Wavelength | 0.979 | 0.979 |
| Space group | P65 | P6522 |
| Cell dimensions | ||
| a, b, c (Å) | 74.61, 74.61, 104.35 | 83.27, 83.27, 141.27 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0–2.18 (2.26–2.18) a | 41.64–2.65 (2.78–2.65) |
| Rmerge b (%) | 20.5 (75.9) | 6.1 (55) |
| I/σ(I) | 13.2 (4.2) | 30.5 (6.5) |
| Completeness (%) | 100 (99.9) | 99.8 (99.6) |
| Redundancy | 19.2 (16.2) | 18.8 (20.1) |
| Refinement | ||
| Resolution (Å) | 37.31–2.18 (2.32–2.18) | 41.64-2.65 (3.03–2.65) |
| No. reflections | 16599 | 8909 |
| Rwork c/Rfree d | 0.183/ 0.225 | 0.254/0.280 |
| No. atoms | ||
| Protein | 1558 | 1746 |
| Ligand (peptide) | 74 | 127 |
| Water | 117 | 0 |
| B-factors (Å2) | ||
| Protein | 31.4 | 81.3 |
| Ligand (peptide) | 45.8 | 90.1 |
| Water | 38.7 | - |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.007 | 0.006 |
| Bond angles (°) | 0.98 | 1.10 |
| Ramachandran favored (%) | 99.06 | 95.90 |
| Allowed (%) | 0.94 | 4.10 |
| Outliers (%) | 0 | 0 |
| MazF-mt9-α4 | MazF-mt1-α3 | ||||
|---|---|---|---|---|---|
| Peptide | MazF-mt9 (Chain A) | MazF-mt9 (Chain B) | Peptide | MazF-mt1 (Chain A) | MazF-mt1 (Chain B) |
| Gly70 (O) | Arg87 (NH2) | Tyr47 (stacking) | Phe61 | ||
| Glu69 | Arg87’ (NH2) | Tyr47 (OH) | His113 (ND1) | Asn31’ (ND2) | |
| Asp74 (OD2) | Ala84 (N) | Glu53 (OE2) | Asn54 (ND2) | ||
| Asp74 (OD1, OD2) | Arg87 (NE, NH2) | Glu53 (OE1) | Lys77(NZ) | ||
| Asp74 (O) | Arg81’(NH1, NH2) | Trp54 (NE1) | Asn54 (ND2) | ||
| Trp54 (stacking) | Asn54 | ||||
| Ser55 (OG) | Arg24 (NH2) | ||||
| Ser55 (O) | Arg24 (NH1) | ||||
| Name | Sequence | Binding Affinity |
|---|---|---|
| E9 α4 (10 residue) | EWEGTVGDGL | No binding |
| E9 α4 | DEDREWEGTVGDGLG | Strong binding |
| E1 α3 | TLEDDYANAWQEWSAAG | Strong binding to MazF-mt1 |
| E9-9 | DEDREAADGTVGDGLG | No binding |
| E9-10 | DEDRAAADGTVGDGLG | No binding |
| E9-12 | DEDRAAADGTDGDGLG | No binding |
| E9-13 | DEDRLWDGTVGDGLG | Strong binding |
| E9-14 | DEDREWDGTVGDWLG | Strong binding |
| E9-15 | DEDREWDGTVGDWERDED | Strong binding |
| E9-16 | GLGDGVTGEWERDED | No binding |
| E9-15AA2 | DEDREWEGDVGDGLG | No binding |
| E9-15AA3 | DEDREWEGTDGDGLG | Strong binding |
| E9-15AA4 | DEDREWDGTVGDGLG | Strong binding |
| E9-15AA5 | DEDREGDGTVGDGLG | No binding |
| E9-15AA6 | DEDRLGDGTVGDGLG | No binding |
| E9-15AA7 | DEDRLGDGTTGDGLG | No binding |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, R.; Zhou, J.; Xie, W. Mechanistic Insight into the Peptide Binding Modes to Two M. tb MazF Toxins. Toxins 2021, 13, 319. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13050319
Chen R, Zhou J, Xie W. Mechanistic Insight into the Peptide Binding Modes to Two M. tb MazF Toxins. Toxins. 2021; 13(5):319. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13050319
Chicago/Turabian StyleChen, Ran, Jie Zhou, and Wei Xie. 2021. "Mechanistic Insight into the Peptide Binding Modes to Two M. tb MazF Toxins" Toxins 13, no. 5: 319. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins13050319