Targeting Programmed Fusobacterium nucleatum Fap2 for Colorectal Cancer Therapy
Abstract
:1. Introduction
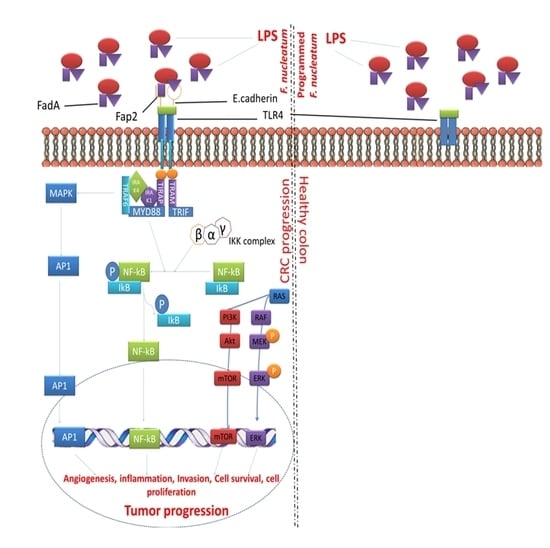
2. F. nucleatum Mediate CRC and Inhibits Host Immune Response
3. Fap2 in F. nucleatum Mediates CRC through Host Gal-GalNAc
4. Bacteria-Mediated Cancer Treatment: Alternative to Surgery
5. Programmed Bacteria
6. Mechanism of Tumoricidal Properties of Programmed Bacteria
7. Targeting Programmed F. nucleatum Fap2 for Colorectal Cancer
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Shang, F.-M.; Liu, H.-L. Fusobacterium nucleatum and colorectal cancer: A review. World J. Gastrointest. Oncol. 2018, 10, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, P.; Yu, Y.; Huang, B.; Zhang, X.; Xu, C.; Zhao, X.; Yin, Z.; He, Z.; Jin, M.; et al. Tumor-preventing activity of aspirin in multiple cancers based on bioinformatic analyses. PeerJ 2018, 6, e5667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, F.; Cai, Y. Study insights into gastrointestinal cancer through the gut microbiota. BioMed Res. Int. 2019, 2019, 8721503. [Google Scholar] [CrossRef] [PubMed]
- Jobin, C. Colorectal cancer: Looking for answers in the microbiota. Cancer Discov. 2013, 3, 384–387. [Google Scholar] [CrossRef] [PubMed]
- Mira-Pascual, L.; Cabrera-Rubio, R.; Ocon, S.; Costales, P.; Parra, A.; Suarez, A.; Moris, F.; Rodrigo, L.; Mira, A.; Collado, M.C. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J. Gastroenterol. 2014, 50, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, K.; Xu, B. Molecular targets of vitexin and isovitexin in cancer therapy: A critical review. Ann. N. Y. Acad. Sci. 2017, 1401, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fu, C.; Zhao, Y.; He, J. Intervention with α-ketoglutarate ameliorates colitis-related colorectal carcinoma via modulation of the gut microbiome. BioMed Res. Int. 2019, 2019, 8020785. [Google Scholar] [CrossRef]
- Ganesan, K.; Chung, S.K.; Vanamala, J.; Xu, B. Causal relationship between diet-induced gut microbiota changes and diabetes: A novel strategy to transplant Faecalibacterium prausnitzii in preventing diabetes. Int. J. Mol. Sci. 2018, 19, 3720. [Google Scholar] [CrossRef]
- Arthur, J.C.; Jobin, C. The complex interplay between inflammation, the microbiota and colorectal cancer. Gut Microbes 2013, 4, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Candela, M. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J. Gastroenterol. 2014, 20, 908. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Belizario, J.E.; Faintuch, J. Microbiome and gut dysbiosis. Exp. Suppl. (2012) 2018, 109, 459–476. [Google Scholar] [CrossRef]
- Viljoen, K.S.; Dakshinamurthy, A.; Goldberg, P.; Blackburn, J.M. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between Fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS ONE 2015, 10, e0119462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; He, H.; Xu, H.; Li, Y.; Li, Z.; Du, Y.; He, J.; Zhou, Y.; Wang, H.; Nie, Y. Association of oncogenic bacteria with colorectal cancer in South China. Oncotarget 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-E.; Chiu, C.-T.; Rayner, C.K.; Wu, K.-L.; Chiu, Y.-C.; Hu, M.-L.; Chuah, S.-K.; Tai, W.-C.; Liang, C.-M.; Wang, H.-M. Associated factors in Streptococcus bovis bacteremia and colorectal cancer. Kaohsiung J. Med. Sci. 2016, 32, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Van Nhieu, J.T.; Furet, J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Eslick, G.D. Streptococcus bovisinfection and colorectal neoplasia: A meta-analysis. Colorectal Dis. 2014, 16, 672–680. [Google Scholar] [CrossRef]
- Gagnière, J. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501. [Google Scholar] [CrossRef]
- Yang, Y.; Misra, B.B.; Liang, L.; Bi, D.; Weng, W.; Wu, W.; Cai, S.; Qin, H.; Goel, A.; Li, X.; et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics 2019, 9, 4101–4114. [Google Scholar] [CrossRef]
- Song, M.; Chan, A.T.; Sun, J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology 2019, in press. [Google Scholar] [CrossRef]
- Yazici, C.; Wolf, P.G.; Kim, H.; Cross, T.L.; Vermillion, K.; Carroll, T.; Augustus, G.J.; Mutlu, E.; Tussing-Humphreys, L.; Braunschweig, C.; et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 2017, 66, 1983–1994. [Google Scholar] [CrossRef]
- Kharrat, N.; Assidi, M.; Abu-Elmagd, M.; Pushparaj, P.N.; Alkhaldy, A.; Arfaoui, L.; Naseer, M.I.; Omri, A.E.; Messaoudi, S.; Buhmeida, A.; et al. Data mining analysis of human gut microbiota links Fusobacterium spp. with colorectal cancer onset. Bioinformation 2019, 15, 372–379. [Google Scholar] [CrossRef]
- Warren, R.L.; Freeman, D.J.; Pleasance, S.; Watson, P.; Moore, R.A.; Cochrane, K.; Allen-Vercoe, E.; Holt, R.A. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013, 1, 16. [Google Scholar] [CrossRef]
- Yu, J.; Feng, Q.; Wong, S.H.; Zhang, D.; Liang, Q.y.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y.; et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2015, 66, 70–78. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2011, 22, 299–306. [Google Scholar] [CrossRef]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatumin colorectal carcinoma tissue and patient prognosis. Gut 2015, 65, 1973–1980. [Google Scholar] [CrossRef]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology 2017, 152, 851–866.e824. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral bacteria and intestinal dysbiosis in colorectal cancer. Int. J. Mol. Sci. 2019, 20, 4146. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver–passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Mark Welch, J.L.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef] [Green Version]
- Marchesi, J.R.; Dutilh, B.E.; Hall, N.; Peters, W.H.M.; Roelofs, R.; Boleij, A.; Tjalsma, H. Towards the human colorectal cancer microbiome. PLoS ONE 2011, 6, e20447. [Google Scholar] [CrossRef]
- Park, C.H.; Han, D.S.; Oh, Y.-H.; Lee, A.R.; Lee, Y.-R.; Eun, C.S. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Komiya, Y.; Shimomura, Y.; Higurashi, T.; Sugi, Y.; Arimoto, J.; Umezawa, S.; Uchiyama, S.; Matsumoto, M.; Nakajima, A. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut 2019, 68, 1335–1337. [Google Scholar] [CrossRef]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Gao, R.; Kong, C.; Huang, L.; Li, H.; Qu, X.; Liu, Z.; Lan, P.; Wang, J.; Qin, H. Mucosa-associated microbiota signature in colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2073–2083. [Google Scholar] [CrossRef]
- Burns, M.B.; Lynch, J.; Starr, T.K.; Knights, D.; Blekhman, R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015, 7. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Amitay, E.L.; Werner, S.; Vital, M.; Pieper, D.H.; Höfler, D.; Gierse, I.-J.; Butt, J.; Balavarca, Y.; Cuk, K.; Brenner, H. Fusobacterium and colorectal cancer: Causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis 2017, 38, 781–788. [Google Scholar] [CrossRef]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef] [PubMed]
- Vogtmann, E.; Hua, X.; Zeller, G.; Sunagawa, S.; Voigt, A.Y.; Hercog, R.; Goedert, J.J.; Shi, J.; Bork, P.; Sinha, R. Colorectal cancer and the human gut microbiome: Reproducibility with whole-genome shotgun sequencing. PLoS ONE 2016, 11, e0155362. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Suehiro, Y.; Hashimoto, S.; Hoshida, T.; Fujimoto, M.; Watanabe, M.; Imanaga, D.; Sakai, K.; Matsumoto, T.; Nishioka, M.; et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J. Gastroenterol. 2018, 53, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Ge, Q.X.; Cao, J.; Zhou, Y.J.; Du, Y.L.; Shen, B.; Wan, Y.J.; Nie, Y.Q. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J. Gastroenterol. 2016, 22, 3227–3233. [Google Scholar] [CrossRef] [PubMed]
- Tahara, T.; Yamamoto, E.; Suzuki, H.; Maruyama, R.; Chung, W.; Garriga, J.; Jelinek, J.; Yamano, H.O.; Sugai, T.; An, B.; et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014, 74, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Suehiro, Y.; Sakai, K.; Nishioka, M.; Hashimoto, S.; Takami, T.; Higaki, S.; Shindo, Y.; Hazama, S.; Oka, M.; Nagano, H.; et al. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann. Clin. Biochem. 2017, 54, 86–91. [Google Scholar] [CrossRef]
- Wong, S.H.; Kwong, T.N.Y.; Chow, T.C.; Luk, A.K.C.; Dai, R.Z.W.; Nakatsu, G.; Lam, T.Y.T.; Zhang, L.; Wu, J.C.Y.; Chan, F.K.L.; et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 2017, 66, 1441–1448. [Google Scholar] [CrossRef]
- Nosho, K.; Sukawa, Y.; Adachi, Y.; Ito, M.; Mitsuhashi, K.; Kurihara, H.; Kanno, S.; Yamamoto, I.; Ishigami, K.; Igarashi, H.; et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J. Gastroenterol. 2016, 22, 557–566. [Google Scholar] [CrossRef]
- Ito, M.; Kanno, S.; Nosho, K.; Sukawa, Y.; Mitsuhashi, K.; Kurihara, H.; Igarashi, H.; Takahashi, T.; Tachibana, M.; Takahashi, H.; et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer 2015, 137, 1258–1268. [Google Scholar] [CrossRef]
- Purcell, R.V.; Visnovska, M.; Biggs, P.J.; Schmeier, S.; Frizelle, F.A. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Allen-Vercoe, E.; Strauss, J.; Chadee, K. Fusobacterium nucleatum. Gut Microbes 2011, 2, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Dorffel, Y.; Loening-Baucke, V.; Theissig, F.; Ruckert, J.C.; Ismail, M.; Rau, W.A.; Gaschler, D.; Weizenegger, M.; Kuhn, S.; et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 2009, 60, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2014, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Irrazábal, T.; Belcheva, A.; Girardin, S.E.; Martin, A.; Philpott, D.J. The multifaceted role of the intestinal microbiota in colon cancer. Mol. Cell 2014, 54, 309–320. [Google Scholar] [CrossRef]
- Yamamura, K.; Baba, Y.; Miyake, K.; Nakamura, K.; Shigaki, H.; Mima, K.; Kurashige, J.; Ishimoto, T.; Iwatsuki, M.; Sakamoto, Y.; et al. Fusobacterium nucleatum in gastroenterological cancer: Evaluation of measurement methods using quantitative polymerase chain reaction and a literature review. Oncol. Lett. 2017, 14, 6373–6378. [Google Scholar] [CrossRef]
- McCoy, A.N.; Araújo-Pérez, F.; Azcárate-Peril, A.; Yeh, J.J.; Sandler, R.S.; Keku, T.O. Fusobacterium is associated with colorectal adenomas. PLoS ONE 2013, 8, e53653. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2011, 22, 292–298. [Google Scholar] [CrossRef]
- Guo, S.-H.; Wang, H.-F.; Nian, Z.-G.; Wang, Y.-D.; Zeng, Q.-Y.; Zhang, G. Immunization with alkyl hydroperoxide reductase subunit C reduces Fusobacterium nucleatum load in the intestinal tract. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Park, H.E.; Kim, J.H.; Cho, N.-Y.; Lee, H.S.; Kang, G.H. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch. 2017, 471, 329–336. [Google Scholar] [CrossRef]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K.; et al. Fusobacterium nucleatumand T cells in colorectal carcinoma. Jama Oncol. 2015, 1, 653. [Google Scholar] [CrossRef] [PubMed]
- Mikuls, T.R.; Payne, J.B.; Yu, F.; Thiele, G.M.; Reynolds, R.J.; Cannon, G.W.; Markt, J.; McGowan, D.; Kerr, G.S.; Redman, R.S.; et al. Periodontitis and Porphyromonas gingivalis in patients with rheumatoid arthritis. Arthritis Rheumatol. 2014, 66, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Velsko, I.M.; Chukkapalli, S.S.; Rivera-Kweh, M.F.; Chen, H.; Zheng, D.; Bhattacharyya, I.; Gangula, P.R.; Lucas, A.R.; Kesavalu, L. Fusobacterium nucleatum alters atherosclerosis risk factors and enhances inflammatory markers with an atheroprotective immune response in ApoE(null) mice. PLoS ONE 2015, 10, e0129795. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; Li, L.F.; Guo, S.H.; Zeng, Q.Y.; Ning, F.; Liu, W.L.; Zhang, G. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci. Rep. 2016, 6, 33440. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Xiao, H.; Guo, S.; Xu, B.; Liao, Y.; Wu, Y.; Zhang, G. Indoleamine 2,3-dioxygenase expression regulates the survival and proliferation of Fusobacterium nucleatum in THP-1-derived macrophages. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Wang, R.; Bhattacharya, R.; Boulbes, D.R.; Fan, F.; Xia, L.; Adoni, H.; Ajami, N.J.; Wong, M.C.; Smith, D.P.; et al. Fusobacterium nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev. Res. 2017, 10, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.; Kaplan, G.G.; Beck, P.L.; Rioux, K.; Panaccione, R.; DeVinney, R.; Lynch, T.; Allen-Vercoe, E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011, 17, 1971–1978. [Google Scholar] [CrossRef]
- Manson McGuire, A.; Cochrane, K.; Griggs, A.D.; Haas, B.J.; Abeel, T.; Zeng, Q.; Nice, J.B.; MacDonald, H.; Birren, B.W.; Berger, B.W.; et al. Evolution of invasion in a diverse set of Fusobacterium species. mBio 2014, 5. [Google Scholar] [CrossRef]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; Horst, R.t.; Jansen, T.; Jacobs, L.; Bonder, M.J.; et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 2016, 167, 1897. [Google Scholar] [CrossRef]
- Karin, M. Nuclear factor-κB in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef]
- DiDonato, J.A.; Mercurio, F.; Karin, M. NF-κB and the link between inflammation and cancer. Immunol. Rev. 2012, 246, 379–400. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, C.W.; Ma, X.; Paranjpe, A.; Jewett, A.; Lux, R.; Kinder-Haake, S.; Shi, W. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect. Immun. 2010, 78, 4773–4778. [Google Scholar] [CrossRef] [PubMed]
- Coppenhagen-Glazer, S.; Sol, A.; Abed, J.; Naor, R.; Zhang, X.; Han, Y.W.; Bachrach, G. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun. 2015, 83, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cai, S.; Ma, Y. Association between Fusobacterium nucleatum and colorectal cancer: Progress and future directions. J. Cancer 2018, 9, 1652–1659. [Google Scholar] [CrossRef]
- Xu, M.; Yamada, M.; Li, M.; Liu, H.; Chen, S.G.; Han, Y.W. Fad A from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J. Biol. Chem. 2007, 282, 25000–25009. [Google Scholar] [CrossRef]
- Santaolalla, R.; Sussman, D.A.; Ruiz, J.R.; Davies, J.M.; Pastorini, C.; España, C.L.; Sotolongo, J.; Burlingame, O.; Bejarano, P.A.; Philip, S.; et al. TLR4 activates the β-catenin pathway to cause intestinal neoplasia. PLoS ONE 2013, 8, e63298. [Google Scholar] [CrossRef]
- Shi, C.; Yang, Y.; Xia, Y.; Okugawa, Y.; Yang, J.; Liang, Y.; Chen, H.; Zhang, P.; Wang, F.; Han, H.; et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut 2015, 65, 1470–1481. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, Y.; Yu, J.; Chen, T.; Wu, Y.; Shi, L.; Li, Q.; Wu, J.; Fu, X. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget 2017, 8. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Guo, S.; Li, L.; Xu, B.; Li, M.; Zeng, Q.; Xiao, H.; Xue, Y.; Wu, Y.; Wang, Y.; Liu, W.; et al. A simple and novel fecal biomarker for colorectal cancer: Ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin. Chem. 2018, 64, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Bashir, A.; Miskeen, A.Y.; Bhat, A.; Fazili, K.M.; Ganai, B.A. Fusobacterium nucleatum. Eur. J. Cancer Prev. 2015, 24, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Hussan, H.; Clinton, S.K.; Roberts, K.; Bailey, M.T. Fusobacterium’s link to colorectal neoplasia sequenced: A systematic review and future insights. World J. Gastroenterol. 2017, 23, 8626–8650. [Google Scholar] [CrossRef] [PubMed]
- Abed, J.; Emgard, J.E.M.; Zamir, G.; Faroja, M.; Almogy, G.; Grenov, A.; Sol, A.; Naor, R.; Pikarsky, E.; Atlan, K.A.; et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 2016, 20, 215–225. [Google Scholar] [CrossRef]
- Abed, J.; Maalouf, N.; Parhi, L.; Chaushu, S.; Mandelboim, O.; Bachrach, G. Tumor targeting by Fusobacterium nucleatum: A pilot study and future perspectives. Front. Cell. Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Rawal, S.; Majumdar, S.; Dhawan, V.; Vohra, H. Entamoeba histolytica Gal/GalNAc lectin depletes antioxidant defences of target epithelial cells. Parasitology 2004, 128, 617–624. [Google Scholar] [CrossRef]
- Sanchez, V.; Serrano-Luna, J.; Ramirez-Moreno, E.; Tsutsumi, V.; Shibayama, M. Entamoeba histolytica: Overexpression of the gal/galnac lectin, ehcp2 and ehcp5 genes in an in vivo model of amebiasis. Parasitol. Int. 2016, 65, 665–667. [Google Scholar] [CrossRef]
- Frederick, J.R.; Petri, W.A., Jr. Roles for the galactose-/N-acetylgalactosamine-binding lectin of Entamoeba in parasite virulence and differentiation. Glycobiology 2005, 15, 53r–59r. [Google Scholar] [CrossRef]
- Burchell, J.M.; Beatson, R.; Graham, R.; Taylor-Papadimitriou, J.; Tajadura-Ortega, V. O-linked mucin-type glycosylation in breast cancer. Biochem. Soc. Trans. 2018, 46, 779–788. [Google Scholar] [CrossRef]
- Tzeng, S.-F.; Tsai, C.-H.; Chao, T.-K.; Chou, Y.-C.; Yang, Y.-C.; Tsai, M.-H.; Cha, T.-L.; Hsiao, P.-W. O-Glycosylation–mediated signaling circuit drives metastatic castration-resistant prostate cancer. FASEB J. 2018, 32, 6869–6882. [Google Scholar] [CrossRef]
- Arend, P. Early ovariectomy reveals the germline encoding of natural anti-A- and Tn-cross-reactive immunoglobulin M (IgM) arising from developmentalO-GalNAc glycosylations. (Germline-encoded natural anti-A/Tn cross-reactive IgM). Cancer Med. 2017, 6, 1601–1613. [Google Scholar] [CrossRef] [PubMed]
- Hung, J.-S.; Huang, J.; Lin, Y.-C.; Huang, M.-J.; Lee, P.-H.; Lai, H.-S.; Liang, J.-T.; Huang, M.-C. C1GALT1 overexpression promotes the invasive behavior of colon cancer cells through modifying O-glycosylation of FGFR2. Oncotarget 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.-Q.; Wan, H.-Y.; Li, H.-F.; Liu, M.; Li, X.; Tang, H. MicroRNA-214 suppresses growth and invasiveness of cervical cancer cells by targeting UDP-N-acetyl-α-d-galactosamine: Polypeptide N-acetylgalactosaminyltransferase 7. J. Biol. Chem. 2012, 287, 14301–14309. [Google Scholar] [CrossRef] [PubMed]
- Chugh, S.; Barkeer, S.; Rachagani, S.; Nimmakayala, R.K.; Perumal, N.; Pothuraju, R.; Atri, P.; Mahapatra, S.; Thapa, I.; Talmon, G.A.; et al. Disruption of C1galt1 gene promotes development and metastasis of pancreatic adenocarcinomas in mice. Gastroenterology 2018, 155, 1608–1624. [Google Scholar] [CrossRef]
- Jiang, X.-N.; Dang, Y.-F.; Gong, F.-L.; Guo, X.-L. Role and regulation mechanism of Gal-3 in non-small cell lung cancer and its potential clinical therapeutic significance. Chem. Biol. Interact. 2019, 309, 108724. [Google Scholar] [CrossRef]
- Tian, Y.; Denda-Nagai, K.; Kamata-Sakurai, M.; Nakamori, S.; Tsukui, T.; Itoh, Y.; Okada, K.; Yi, Y.; Irimura, T. Mucin 21 in esophageal squamous epithelia and carcinomas: Analysis with glycoform-specific monoclonal antibodies. Glycobiology 2012, 22, 1218–1226. [Google Scholar] [CrossRef]
- Han, Y.W.; Ikegami, A.; Rajanna, C.; Kawsar, H.I.; Zhou, Y.; Li, M.; Sojar, H.T.; Genco, R.J.; Kuramitsu, H.K.; Deng, C.X. Identification and characterization of a novel adhesin unique to oral Fusobacteria. J. Bacteriol. 2005, 187, 5330–5340. [Google Scholar] [CrossRef]
- Wu, J.; Li, Q.; Fu, X. Fusobacterium nucleatum contributes to the carcinogenesis of colorectal cancer by inducing inflammation and suppressing host immunity. Transl. Oncol. 2019, 12, 846–851. [Google Scholar] [CrossRef]
- Ashare, A.; Stanford, C.; Hancock, P.; Stark, D.; Lilli, K.; Birrer, E.; Nymon, A.; Doerschug, K.C.; Hunninghake, G.W. Chronic liver disease impairs bacterial clearance in a human model of induced Bacteremia. Clin. Transl. Sci. 2009, 2, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Ghidini, M.; Petrillo, A.; Salati, M.; Khakoo, S.; Varricchio, A.; Tomasello, G.; Grossi, F.; Petrelli, F. Surgery or locoregional approaches for hepatic oligometastatic pancreatic cancer: Myth, hope, or reality? Cancers 2019, 11, 1095. [Google Scholar] [CrossRef]
- Spałek, M.; Michalski, K.; Bujko, K.; Wyrwicz, L. Association between preoperative pelvic irradiation and toxicity of subsequent chemotherapy in rectal cancer. Oncol. Res. Treat. 2019, 42, 497–504. [Google Scholar] [CrossRef]
- Mitchell, D.; Puckett, Y.; Nguyen, Q.N. Literature review of current management of colorectal liver metastasis. Cureus 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Garrett, W.S. Gut Microbiota, inflammation, and colorectal cancer. Annu. Rev. Microbiol. 2016, 70, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.H.; Min, J.-J. Salmonella-mediated cancer therapy: Roles and potential. Nucl. Med. Mol. Imaging 2016, 51, 118–126. [Google Scholar] [CrossRef]
- Chowdhury, S.; Castro, S.; Coker, C.; Hinchliffe, T.E.; Arpaia, N.; Danino, T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 2019, 25, 1057–1063. [Google Scholar] [CrossRef]
- Dougan, M.; Dougan, S.K. Programmable bacteria as cancer therapy. Nat. Med. 2019, 25, 1030–1031. [Google Scholar] [CrossRef]
- Wu, M.-R.; Jusiak, B.; Lu, T.K. Engineering advanced cancer therapies with synthetic biology. Nat. Rev. Cancer 2019, 19, 187–195. [Google Scholar] [CrossRef]
- Chien, T.; Doshi, A.; Danino, T. Advances in bacterial cancer therapies using synthetic biology. Curr. Opin. Syst. Biol. 2017, 5, 1–8. [Google Scholar] [CrossRef]
- Lim, W.A.; June, C.H. The principles of engineering immune cells to treat cancer. Cell 2017, 168, 724–740. [Google Scholar] [CrossRef]
- Harimoto, T.; Singer, Z.S.; Velazquez, O.S.; Zhang, J.; Castro, S.; Hinchliffe, T.E.; Mather, W.; Danino, T. Rapid Screening of Engineered Microbial Therapies in a 3-D Multicellular Model; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2018. [Google Scholar] [CrossRef]
- Yoon, W.; Yoo, Y.; Chae, Y.S.; Kee, S.H.; Kim, B.M. Therapeutic advantage of genetically engineered Salmonella typhimurium carrying short hairpin RNA against inhibin alpha subunit in cancer treatment. Ann. Oncol. 2018, 29, 2010–2017. [Google Scholar] [CrossRef]
- Panthel, K.; Meinel, K.M.; Sevil Domènech, V.E.; Geginat, G.; Linkemann, K.; Busch, D.H.; Rüssmann, H. Prophylactic anti-tumor immunity against a murine fibrosarcoma triggered by the Salmonella type III secretion system. Microbes Infect. 2006, 8, 2539–2546. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.H.; Kim, H.S.; Ha, J.M.; Hong, Y.; Choy, H.E.; Min, J.J. Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res. 2009, 70, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.M.; Green, J.; Williams, P.J.; Tazzyman, S.; Hunt, S.; Harmey, J.H.; Kehoe, S.C.; Lewis, C.E. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther. 2009, 16, 329–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loeffler, M.; Le’Negrate, G.; Krajewska, M.; Reed, J.C. Salmonella typhimurium engineered to produce CCL21 inhibit tumor growth. Cancer Immunol. Immunother. 2008, 58, 769–775. [Google Scholar] [CrossRef]
- Loeffler, M.; Le’Negrate, G.; Krajewska, M.; Reed, J.C. Attenuated Salmonella engineered to produce human cytokine LIGHT inhibit tumor growth. Proc. Natl. Acad. Sci. USA 2007, 104, 12879–12883. [Google Scholar] [CrossRef]
- Xiang, R.; Mizutani, N.; Luo, Y.; Chiodoni, C.; Zhou, H.; Mizutani, M.; Ba, Y.; Becker, J.C.; Reisfeld, R.A. A DNA vaccine targeting survivin combines apoptosis with suppression of angiogenesis in lung tumor eradication. Cancer Res. 2005, 65, 553–561. [Google Scholar]
- Agorio, C.; Schreiber, F.; Sheppard, M.; Mastroeni, P.; Fernandez, M.; Martinez, M.A.; Chabalgoity, J.A. Live attenuated Salmonella as a vector for oral cytokine gene therapy in melanoma. J. Gene Med. 2007, 9, 416–423. [Google Scholar] [CrossRef]
- Wen, L.J.; Gao, L.F.; Jin, C.S.; Zhang, H.J.; Ji, K.; Yang, J.P.; Zhao, X.J.; Wen, M.J.; Guan, G.F. Small interfering RNA survivin and GRIM-19 co-expression salmonella plasmid inhibited the growth of laryngeal cancer cells in vitro and in vivo. Int. J. Clin. Exp. Pathol. 2013, 6, 2071–2081. [Google Scholar]
- Liu, Y.-B.; Zhang, L.; Guo, Y.-X.; Gao, L.-F.; Liu, X.-C.; Zhao, L.-J.; Guo, B.-F.; Zhao, L.-J.; Zhao, X.-J.; Xu, D.-Q. Plasmid-based Survivin shRNA and GRIM-19 carried by attenuated Salmonella suppresses tumor cell growth. Asian J. Androl. 2012, 14, 536–545. [Google Scholar] [CrossRef] [Green Version]
- Li, Z. Anti-angiogenesis and anticancer effects of a plasmid expressing both ENDO-VEGI151 and small interfering RNA against survivin. Int. J. Mol. Med. 2011, 29, 485–490. [Google Scholar] [CrossRef]
- Nishikawa, H. In vivo antigen delivery by a Salmonella typhimurium type III secretion system for therapeutic cancer vaccines. J. Clin. Investig. 2006, 116, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, M.; Le’Negrate, G.; Krajewska, M.; Reed, J.C. Inhibition of tumor growth using Salmonella expressing Fas ligand. JNCI J. Natl. Cancer Inst. 2008, 100, 1113–1116. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Chu, L.; Han, X.; Liu, X.; Ren, D. Synergistic antitumoral effects of human telomerase reverse transcriptase-mediated dual-apoptosis-related gene vector delivered by orally attenuated Salmonella enterica Serovar Typhimurium in murine tumor models. J. Gene Med. 2008, 10, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.-S.; Nitcheu-Tefit, J.; Alcock, S.; Ramirez-Jimenez, F.; Chao, T.-Y.; Baril, P.; Rocha, M.; Brett, S.J.; Stauss, H.J.; Vassaux, G. Development of an Escherichia coli expressing Listeriolysin-O vaccine against wilms tumor gene 1-expressing tumors. J. Immunother. 2009, 32, 845–855. [Google Scholar] [CrossRef]
- Yang, N.; Zhu, X.; Chen, L.; Li, S.; Ren, D. Oral administration of attenuated S. typhimurium carrying shRNA-expressing vectors as a cancer therapeutic. Cancer Biol. Ther. 2008, 7, 145–151. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Hu, J.; Wang, B.; Zhao, L.; Ji, K.; Guo, B.; Yin, D.; Du, Y.; Kopecko, D.J.; et al. Plasmid-based E6-specific siRNA and co-expression of wild-type p53 suppresses the growth of cervical cancer in vitro and in vivo. Cancer Lett. 2013, 335, 242–250. [Google Scholar] [CrossRef] [Green Version]
- Sorenson, B.S.; Banton, K.L.; Frykman, N.L.; Leonard, A.S.; Saltzman, D.A. Attenuated Salmonella typhimurium with interleukin 2 gene prevents the establishment of pulmonary metastases in a model of osteosarcoma. J. Pediatric Surg. 2008, 43, 1153–1158. [Google Scholar] [CrossRef]
- Jiang, T.; Zhou, C.; Gu, J.; Liu, Y.; Zhao, L.; Li, W.; Wang, G.; Li, Y.; Cai, L. Enhanced therapeutic effect of cisplatin on the prostate cancer in tumor-bearing mice by transfecting the attenuated Salmonella carrying a plasmid co-expressing p53 gene and mdm2 siRNA. Cancer Lett. 2013, 337, 133–142. [Google Scholar] [CrossRef]
- Ganai, S.; Arenas, R.B.; Forbes, N.S. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br. J. Cancer 2009, 101, 1683–1691. [Google Scholar] [CrossRef]
- Flentie, K.; Kocher, B.; Gammon, S.T.; Novack, D.V.; McKinney, J.S.; Piwnica-Worms, D. A Bioluminescent transposon reporter-trap identifies tumor-specific microenvironment-induced promoters in Salmonella for conditional bacterial-based tumor therapy. Cancer Discov. 2012, 2, 624–637. [Google Scholar] [CrossRef]
- Tian, Y.; Guo, B.; Jia, H.; Ji, K.; Sun, Y.; Li, Y.; Zhao, T.; Gao, L.; Meng, Y.; Kalvakolanu, D.V.; et al. Targeted therapy via oral administration of attenuated Salmonella expression plasmid-vectored Stat3-shRNA cures orthotopically transplanted mouse HCC. Cancer Gene Ther. 2012, 19, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gao, L.; Zhao, L.; Guo, B.; Ji, K.; Tian, Y.; Wang, J.; Yu, H.; Hu, J.; Kalvakolanu, D.V.; et al. Intratumoral delivery and suppression of prostate tumor growth by attenuated Salmonella enterica serovar typhimurium carrying plasmid-based small interfering RNAs. Cancer Res. 2007, 67, 5859–5864. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Wang, B.; Ji, K.; Liang, Z.; Guo, B.; Hu, J.; Yin, D.; Du, Y.; Kopecko, D.J.; et al. Delivery of the co-expression plasmid pEndo-Si-Stat3 by attenuated Salmonella serovar typhimurium for prostate cancer treatment. J. Cancer Res. Clin. Oncol. 2013, 139, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Fensterle, J.; Bergmann, B.; Yone, C.L.R.P.; Hotz, C.; Meyer, S.R.; Spreng, S.; Goebel, W.; Rapp, U.R.; Gentschev, I. Cancer immunotherapy based on recombinant Salmonella enterica serovar Typhimurium aroA strains secreting prostate-specific antigen and cholera toxin subunit B. Cancer Gene Ther. 2007, 15, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, M.; Le’Negrate, G.; Krajewska, M.; Reed, J.C. IL-18-producing Salmonella inhibit tumor growth. Cancer Gene Ther. 2008, 15, 787–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niethammer, A.G.; Xiang, R.; Becker, J.C.; Wodrich, H.; Pertl, U.; Karsten, G.; Eliceiri, B.P.; Reisfeld, R.A. A DNA vaccine against VEGF receptor 2 prevents effective angiogenesis and inhibits tumor growth. Nat. Med. 2002, 8, 1369–1375. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial secretion systems: An overview. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Wu, X.Y.; Chapman, T.; Trott, D.J.; Bettelheim, K.; Do, T.N.; Driesen, S.; Walker, M.J.; Chin, J. Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl. Environ. Microbiol. 2007, 73, 83–91. [Google Scholar] [CrossRef]
- Luo, X.; Li, Z.; Lin, S.; Le, T.; Ittensohn, M.; Bermudes, D.; Runyab, J.D.; Shen, S.y.; Chen, J.; King, I.C.; et al. Antitumor effect of VNP20009, an attenuated Salmonella, in murine tumor models. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2001, 12, 501–508. [Google Scholar] [CrossRef]
- Sznol, M.; Lin, S.L.; Bermudes, D.; Zheng, L.-m.; King, I. Use of preferentially replicating bacteria for the treatment of cancer. J. Clin. Investig. 2000, 105, 1027–1030. [Google Scholar] [CrossRef] [Green Version]
- Hashemi Goradel, N.; Heidarzadeh, S.; Jahangiri, S.; Farhood, B.; Mortezaee, K.; Khanlarkhani, N.; Negahdari, B. Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J. Cell. Physiol. 2018, 234, 2337–2344. [Google Scholar] [CrossRef] [PubMed]


| Sample Tested | Total Number of Clinical Samples | Detection Method | Positive Percentage | References |
|---|---|---|---|---|
| FFPE tissue | 6 | 16S rRNA | 32 | [32] |
| FFPE tissue | 6 | 16S rRNA | 100 | [33] |
| FFPE tissue | 8 | 16S rRNA | 100 | [34] |
| Feces | 14 | qPCR | 57 | [35] |
| FFPE tissue | 31 | 16S rRNA | 10 | [36] |
| FFPE tissue | 37 | 16S rRNA | 9 | [37] |
| FFPE tissue | 44 | 16S rRNA | 100 | [38] |
| FFPE tissue | 46 | 16S rRNA | 100 | [39] |
| FFPE tissue | 46 | 16S rRNA | 54 | [40] |
| FFPE tissue | 47 | 16S rRNA | 32 | [41] |
| FFPE tissue | 52 | 16S rRNA | 77 | [42] |
| Feces | 72 | qPCR | 64 | [43] |
| FFPE tissue | 97 | 16S rRNA | 72 | [14] |
| Frozen tissue and FFPE tissue | 101 | FISH and FQ-PCR | 87 | [44] |
| Genomic DNA | 149 | qPCR | 74 | [45] |
| Feces | 158 | ddPCR | 54 | [46] |
| FFPE tissue | 309 | qPCR | 34 | [47] |
| FFPE tissue | 511 | qPCR | 9 | [48] |
| FFPE tissue | 504 | qPCR | 56 | [49] |
| FFPE tissue | 598 | qPCR | 13 | [26] |
| Bacterial Species | Agent(s) | Host | Origin of the Tumor | Tumor(s) | Effector(s) | Results | References |
|---|---|---|---|---|---|---|---|
| S. typhimurium | Bacterial antigen S. typhimurium secreting L. monocytogenes Iap217–225 (Lm-p60) | BALB/c | Bones | WEHI-164 (Fibrosarcoma) cells expressing Lm-p60 | CD8+ cell-mediated | Antigen-specific tumor inhibition | [112] |
| S. typhimurium | Bacterial toxin S. typhimurium secreting HlyE | BALB/c | Breast | CT-26, 4T1 | Not reported | Reduction in tumor mass | [113,114] |
| S. typhimurium | Birc5 (Survivin) | C57BL/6 | Lungs | D121 | CD8+ cell-mediated | Suppression of angiogenesis and pulmonary metastasized tumors | [117] |
| S. typhimurium | BIRC5 shRNA NDUFA13 (GRIM-19) | Nude Mice | Larynx, prostate | Hep-2 (Laryngeal cancer) DU145 (PC-Xenograft) | Apoptosis | Tumor growth reduced | [119,120] |
| S. typhimurium | BIRC5 shRNA TNFSF15 (VEGI) | Nude mice | Breast | MDA-MB-231 (BC-Xenograft) | Apoptosis | Tumor growth reduced | [121] |
| S. typhimurium | ccl21 | BALB/C | Breast | D2F2, CT-26 | CD4+ and CD8+ cell mediated | Tumor-limited inflammatory reaction with a substantial reduction in tumor burden | [115] |
| S. typhimurium | CTAG1B (NY-ESO-1) | BALB/c | Skin | CMS5 cells expressing human NY-ESO-1 | CD8+ cell-mediated | NY-ESO-1-positive tumors are eliminated | [122] |
| S. typhimurium | Cytokine ccl21 | C57BL/6 | Lungs | D121 (LC-Syngeneic) | CD8+ cell mediated | Suppression of angiogenesis and growth of pulmonary metastasized tumors | [117] |
| S. typhimurium | Death inducer S. typhimurium secreting murine Fasl | BALB/c | Breast, colon | CT-26 D2F2 (BC-Syngeneic) | Neutrophils | Reduction in tumor mass | [123] |
| S. typhimurium | Diablo/Trail | BALB/c C57BL/6 | Liver, spleen, kidney | 4T1 LL/2 (LC-Syngeneic) B16F10 (Melanoma) | Apoptosis | Tumor growth inhibition with prolonged survival | [124] |
| E. coli | E. coli expressing LLO | C57BL/6 | Blood | MBL2 (Leukemia-Syngeneic) TRAMP-C (PC-Syngeneic) | CD8+ cell-mediated | Reduction in tumor mass | [125] |
| S. typhimurium | Growth inhibitor(s) Bcl2 shRNA | C57BL/6 | Liver, spleen, skin | B16F10 | Apoptosis | Survival time of tumor-bearing mice Prolonged Complete tumor regression not observed | [126] |
| S. typhimurium | HPV E6 shRNA TP53 | Nude mice | Cervix | SiHa | Apoptosis | Tumor growth reduced | [127] |
| S. typhimurium | IL 18 | C57BL/6 | Skin | B16F1A (Melanoma) | Not reported | Increased survival time | [118] |
| BALB/C | Skin, colon | D2F2, CT-26 | Granulocyte, NK, CD4+, CD8+ cell mediated | Reduced tumor growth and pulmonary metastases | [111] | ||
| S. typhimurium | IL2 | C57BL/6 | Liver | MCA-38 (Adenocarcinoma Syngeneic) | NK cells | Hepatic metastases reduced | [116] |
| BALB/C | Bone, lungs | K7M2 (Osteosarcoma –Syngeneic) | NK cells | Pulmonary metastases reduced compared to saline control | [128] | ||
| S. typhimurium | IL4 | C57BL/6 | Skin | B16F1A (Melanoma) | Not reported | Increased survival time | [118] |
| MDM2 shRNA TP53-Cisplatin | Nude Mice | Prostate | PC3 | Apoptosis | Tumor growth reduced | [129] | |
| S. typhimurium | S. typhimurium secreting murine Trail | BALB/c | Breast | 4T1 | Apoptosis | Tumor growth reduced | [130] |
| S. typhimurium | S. typhimurium secreting Stx2 | Nude Mice | Skin, colon | B16, HCT116, HeLa | Necrosis | Reduction in tumor mass | [131] |
| S. typhimurium | Stat3 shRNA | C57BL/6 | Bone | H22 | Apoptosis and CD8+ cell mediated | Tumor growth reduced | [132] |
| S. typhimurium | Stat3 shRNA Col18A1Endo | C57BL/6 | Prostate | RM1 | Apoptosis | Tumor growth reduced | [133,134] |
| S. typhimurium | Target antigen KLK3 (PSA) | DBA/2 | Prostate | P815 cells expressing human PSA | CD8+ cell-mediated | Direct i.m. DNA vaccination was better than Serovar typhimurium-delivered immunogen | [135] |
| S. typhimurium | Tnfsf14 (LIGHT) | BALB/C | Breast | D2F2, CT-26 | NK, CD4+, CD8+ cell- mediated | Primary and metastatic tumor growth inhibited | [136] |
| S. typhimurium | Vegfr2 (Kdr or Flk1) full-length Protein | BALB/c C57BL/6 | Skin | CT-26 B16G3.26 (Melanoma) D121 MC-38 (CRC-Syngeneic) | CD8+ cell-mediated | Microvessel destruction retarded tumor growth and metastases. Healing of skin wounds slightly delayed. Immunological memory persisted at 120 days post-immunization | [137] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganesan, K.; Guo, S.; Fayyaz, S.; Zhang, G.; Xu, B. Targeting Programmed Fusobacterium nucleatum Fap2 for Colorectal Cancer Therapy. Cancers 2019, 11, 1592. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11101592
Ganesan K, Guo S, Fayyaz S, Zhang G, Xu B. Targeting Programmed Fusobacterium nucleatum Fap2 for Colorectal Cancer Therapy. Cancers. 2019; 11(10):1592. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11101592
Chicago/Turabian StyleGanesan, Kumar, Songhe Guo, Sundaz Fayyaz, Ge Zhang, and Baojun Xu. 2019. "Targeting Programmed Fusobacterium nucleatum Fap2 for Colorectal Cancer Therapy" Cancers 11, no. 10: 1592. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11101592