Inhibitor of DNA-Binding Protein 4 Suppresses Cancer Metastasis through the Regulation of Epithelial Mesenchymal Transition in Lung Adenocarcinoma
Abstract
:1. Introduction
2. Results
2.1. Id4 Expression Inversely Correlates with Lung Cancer Cell Invasiveness
2.2. Expression of Id4 could Interfere with the Malignant Behavior of Lung Cancer Cells In Vitro and In Vivo
2.3. Id4 Affects the Malignancy of Cancer Cell through the Regulation of Epithelial–Mesenchymal Transition
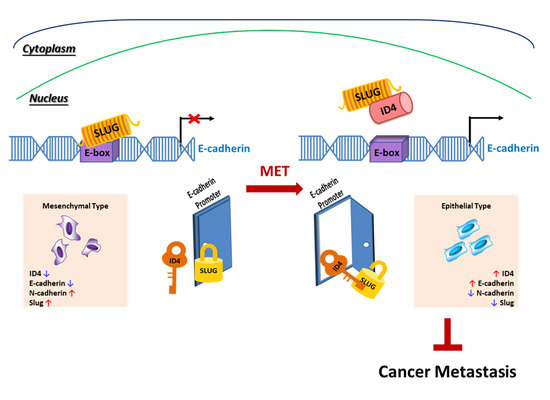
2.4. Id4 Promotes E-cad Expression through the Binding of Slug
2.5. Low-Level Id4 Expressions Are Associated with A Poor Clinical Outcome for LADC Patients
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture Condition
4.2. Reverse Transcription Polymerase Chain Reaction (RT-PCR) and Microarray Analysis
4.3. Plasmid Constructs and Stable Cell Selection
4.4. Short Hairpin RNA (shRNA) and Lentiviral Infection
4.5. Immunoprecipitation and Immunoblotting
4.6. Cell Proliferation and Apoptosis
4.7. Modified Boyden Chamber Invasion Assay
4.8. Luciferase Reporter Assay
4.9. Chromatin Immunoprecipitation Assay (ChIP)
4.10. Experimental Metastasis In Vivo
4.11. IHC Analysis of Tumor Samples from the Lungs of Mice
4.12. TUNEL Assay
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reck, M.; Rabe, K.F. Precision Diagnosis and Treatment for Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 849–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borczuk, A.C. Prognostic considerations of the new World Health Organization classification of lung adenocarcinoma. Eur. Respir. Rev. 2016, 25, 364–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valastyan, S.; Reinhardt, F.; Benaich, N.; Calogrias, D.; Szasz, A.M.; Wang, Z.C.; Brock, J.E.; Richardson, A.L.; Weinberg, R.A. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009, 137, 1032–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steeg, P.S. Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med. 2006, 12, 895–904. [Google Scholar] [CrossRef]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef]
- Pan, S.H.; Chao, Y.C.; Hung, P.F.; Chen, H.Y.; Yang, S.C.; Chang, Y.L.; Wu, C.T.; Chang, C.C.; Wang, W.L.; Chan, W.K.; et al. The ability of LCRMP-1 to promote cancer invasion by enhancing filopodia formation is antagonized by CRMP-1. J. Clin. Investig. 2011, 121, 3189–3205. [Google Scholar] [CrossRef] [Green Version]
- Steeg, P.S. Metastasis suppressors alter the signal transduction of cancer cells. Nat. Rev. Cancer 2003, 3, 55–63. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. Emt: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, M.C.; Gonzalez, S.F.; Welin, J.; Fuxe, J. Epithelial-mesenchymal transition in cancer metastasis through the lymphatic system. Mol. Oncol. 2017, 11, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.; Yoon, C.H.; Kim, R.K.; Lim, E.J.; Oh, Y.S.; Hwang, S.G.; An, S.; Yoon, G.; Gye, M.C.; Yi, J.M.; et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene 2013, 32, 4873–4882. [Google Scholar] [CrossRef] [PubMed]
- Shirkoohi, R. Epithelial mesenchymal transition from a natural gestational orchestration to a bizarre cancer disturbance. Cancer Sci. 2013, 104, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Li, C.L.; Yang, D.; Cao, X.; Wang, F.; Hong, D.Y.; Wang, J.; Shen, X.C.; Chen, Y. Fibronectin induces epithelial-mesenchymal transition in human breast cancer MCF-7 cells via activation of calpain. Oncol. Lett. 2017, 13, 3889–3895. [Google Scholar] [CrossRef] [Green Version]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Wheelock, M.J.; Shintani, Y.; Maeda, M.; Fukumoto, Y.; Johnson, K.R. Cadherin switching. J. Cell Sci. 2008, 121 Pt 6, 727–735. [Google Scholar] [CrossRef] [Green Version]
- Yokota, Y.; Mori, S. Role of Id family proteins in growth control. J. Cell. Physiol. 2002, 190, 21–28. [Google Scholar] [CrossRef]
- Perk, J.; Iavarone, A.; Benezra, R. Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer 2005, 5, 603–614. [Google Scholar] [CrossRef]
- Ruzinova, M.B.; Benezra, R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003, 13, 410–418. [Google Scholar] [CrossRef]
- Norton, J.D. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J. Cell Sci. 2000, 113 Pt 22, 3897–3905. [Google Scholar]
- Lasorella, A.; Uo, T.; Iavarone, A. Id proteins at the cross-road of development and cancer. Oncogene 2001, 20, 8326–8333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagliuca, A.; Bartoli, P.C.; Saccone, S.; Della Valle, G.; Lania, L. Molecular cloning of ID4, a novel dominant negative helix-loop-helix human gene on chromosome 6p21.3–p22. Genomics 1995, 27, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Kamalian, L.; Gosney, J.R.; Forootan, S.S.; Foster, C.S.; Bao, Z.Z.; Beesley, C.; Ke, Y. Increased expression of Id family proteins in small cell lung cancer and its prognostic significance. Clin. Cancer Res. 2008, 14, 2318–2325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, E.C.; Deed, R.W.; Inoue, T.; Norton, J.D.; Sharrocks, A.D. Id helix-loop-helix proteins antagonize pax transcription factor activity by inhibiting DNA binding. Mol. Cell. Biol. 2001, 21, 524–533. [Google Scholar] [CrossRef] [Green Version]
- Moldes, M.; Boizard, M.; Liepvre, X.L.; Feve, B.; Dugail, I.; Pairault, J. Functional antagonism between inhibitor of DNA binding (Id) and adipocyte determination and differentiation factor 1/sterol regulatory element-binding protein-1c (ADD1/SREBP-1c) trans-factors for the regulation of fatty acid synthase promoter in adipocytes. Biochem. J. 1999, 344 Pt 3, 873–880. [Google Scholar] [CrossRef]
- Shoji, W.; Inoue, T.; Yamamoto, T.; Obinata, M. MIDA1, a protein associated with Id, regulates cell growth. J. Biol. Chem. 1995, 270, 24818–24825. [Google Scholar] [CrossRef] [Green Version]
- Ohtani, N.; Zebedee, Z.; Huot, T.J.; Stinson, J.A.; Sugimoto, M.; Ohashi, Y.; Sharrocks, A.D.; Peters, G.; Hara, E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 2001, 409, 1067–1070. [Google Scholar] [CrossRef]
- Yates, P.R.; Atherton, G.T.; Deed, R.W.; Norton, J.D.; Sharrocks, A.D. Id helix-loop-helix proteins inhibit nucleoprotein complex formation by the TCF ETS-domain transcription factors. EMBO J. 1999, 18, 968–976. [Google Scholar] [CrossRef]
- Lasorella, A.; Iavarone, A.; Israel, M.A. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol. Cell. Biol. 1996, 16, 2570–2578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iavarone, A.; Garg, P.; Lasorella, A.; Hsu, J.; Israel, M.A. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994, 8, 1270–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deed, R.W.; Armitage, S.; Norton, J.D. Nuclear localization and regulation of Id protein through an E protein-mediated chaperone mechanism. J. Biol. Chem. 1996, 271, 23603–23606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasskarl, J.; Munger, K. Id proteins--tumor markers or oncogenes? Cancer Biol. Ther. 2002, 1, 91–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sikder, H.A.; Devlin, M.K.; Dunlap, S.; Ryu, B.; Alani, R.M. Id proteins in cell growth and tumorigenesis. Cancer Cell 2003, 3, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Vandeputte, D.A.; Troost, D.; Leenstra, S.; Ijlst-Keizers, H.; Ramkema, M.; Bosch, D.A.; Baas, F.; Das, N.K.; Aronica, E. Expression and distribution of id helix-loop-helix proteins in human astrocytic tumors. Glia 2002, 38, 329–338. [Google Scholar] [CrossRef]
- Ouyang, X.S.; Wang, X.; Lee, D.T.; Tsao, S.W.; Wong, Y.C. Over expression of ID-1 in prostate cancer. J. Urol. 2002, 167, 2598–2602. [Google Scholar] [CrossRef]
- Morrow, M.A.; Mayer, E.W.; Perez, C.A.; Adlam, M.; Siu, G. Overexpression of the Helix-Loop-Helix protein Id2 blocks T cell development at multiple stages. Mol. Immunol. 1999, 36, 491–503. [Google Scholar] [CrossRef]
- Dell′Orso, S.; Ganci, F.; Strano, S.; Blandino, G.; Fontemaggi, G. ID4: A new player in the cancer arena. Oncotarget 2010, 1, 48–58. [Google Scholar] [CrossRef] [Green Version]
- Nasif, D.; Campoy, E.; Laurito, S.; Branham, R.; Urrutia, G.; Roque, M.; Branham, M.T. Epigenetic regulation of ID4 in breast cancer: Tumor suppressor or oncogene? Clin. Epigenet. 2018, 10, e111. [Google Scholar] [CrossRef]
- Yuen, H.F.; Chua, C.W.; Chan, Y.P.; Wong, Y.C.; Wang, X.; Chan, K.W. Id proteins expression in prostate cancer: High-level expression of Id-4 in primary prostate cancer is associated with development of metastases. Mod. Pathol. 2006, 19, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Carey, J.P.; Asirvatham, A.J.; Galm, O.; Ghogomu, T.A.; Chaudhary, J. Inhibitor of differentiation 4 (Id4) is a potential tumor suppressor in prostate cancer. BMC Cancer 2009, 9, e173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontemaggi, G.; Dell′Orso, S.; Trisciuoglio, D.; Shay, T.; Melucci, E.; Fazi, F.; Terrenato, I.; Mottolese, M.; Muti, P.; Domany, E.; et al. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat. Struct. Mol. Biol. 2009, 16, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Rahme, G.J.; Israel, M.A. Id4 suppresses MMP2-mediated invasion of glioblastoma-derived cells by direct inactivation of Twist1 function. Oncogene 2015, 34, 53–62. [Google Scholar] [CrossRef]
- Crippa, E.; Lusa, L.; De Cecco, L.; Marchesi, E.; Calin, G.A.; Radice, P.; Manoukian, S.; Peissel, B.; Daidone, M.G.; Gariboldi, M.; et al. miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PLoS ONE 2014, 9, e87039. [Google Scholar] [CrossRef]
- Chen, J.J.; Peck, K.; Hong, T.M.; Yang, S.C.; Sher, Y.P.; Shih, J.Y.; Wu, R.; Cheng, J.L.; Roffler, S.R.; Wu, C.W.; et al. Global analysis of gene expression in invasion by a lung cancer model. Cancer Res. 2001, 61, 5223–5230. [Google Scholar]
- Chu, Y.W.; Yang, P.C.; Yang, S.C.; Shyu, Y.C.; Hendrix, M.J.; Wu, R.; Wu, C.W. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am. J. Respir. Cell Mol. Biol. 1997, 17, 353–360. [Google Scholar] [CrossRef] [Green Version]
- Hemavathy, K.; Guru, S.C.; Harris, J.; Chen, J.D.; Ip, Y.T. Human Slug is a repressor that localizes to sites of active transcription. Mol. Cell. Biol. 2000, 20, 5087–5095. [Google Scholar] [CrossRef] [Green Version]
- Bolos, V.; Peinado, H.; Perez-Moreno, M.A.; Fraga, M.F.; Esteller, M.; Cano, A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: A comparison with Snail and E47 repressors. J. Cell Sci. 2003, 116 Pt 3, 499–511. [Google Scholar] [CrossRef] [Green Version]
- Hajra, K.M.; Chen, D.Y.; Fearon, E.R. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002, 62, 1613–1618. [Google Scholar]
- Yamauchi, M.; Yamaguchi, R.; Nakata, A.; Kohno, T.; Nagasaki, M.; Shimamura, T.; Imoto, S.; Saito, A.; Ueno, K.; Hatanaka, Y.; et al. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS ONE 2012, 7, e43923. [Google Scholar] [CrossRef] [PubMed]
- Amaral, L.H.P.; Bufalo, N.E.; Peres, K.C.; Barreto, I.S.; Campos, A.; Ward, L.S. ID Proteins May Reduce Aggressiveness of Thyroid Tumors. Endocr. Pathol. 2019, 30, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Fan, J.; Ma, Y.; Zhou, Y.; Qin, K.; Shi, M.; Yang, J. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019, 9, e28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.P.; Wang, W.L.; Chang, Y.L.; Wu, C.T.; Chao, Y.C.; Kao, S.H.; Yuan, A.; Lin, C.W.; Yang, S.C.; Chan, W.K.; et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat. Cell Biol. 2009, 11, 694–704. [Google Scholar] [CrossRef]
- Hung, P.F.; Hong, T.M.; Chang, C.C.; Hung, C.L.; Hsu, Y.L.; Chang, Y.L.; Wu, C.T.; Chang, G.C.; Chan, N.L.; Yu, S.L.; et al. Hypoxia-induced Slug SUMOylation enhances lung cancer metastasis. J. Exp. Clin. Cancer Res. 2019, 38, e5. [Google Scholar] [CrossRef]
- Shih, J.Y.; Yang, P.C. The EMT regulator slug and lung carcinogenesis. Carcinogenesis 2011, 32, 1299–1304. [Google Scholar] [CrossRef] [Green Version]
- Nieto, M.A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002, 3, 155–166. [Google Scholar] [CrossRef]
- Teachenor, R.; Beck, K.; Wright, L.Y.; Shen, Z.; Briggs, S.P.; Murre, C. Biochemical and phosphoproteomic analysis of the helix-loop-helix protein E47. Mol. Cell. Biol. 2012, 32, 1671–1682. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.S.; Yeom, S.; Kwak, J.; Ahn, H.J.; Jang, K.L. Hepatitis B virus X protein induces epithelial-mesenchymal transition by repressing E-cadherin expression via upregulation of E12/E47. J. Gen. Virol. 2016, 97, 134–143. [Google Scholar]
- Pan, S.H.; Chao, Y.C.; Chen, H.Y.; Hung, P.F.; Lin, P.Y.; Lin, C.W.; Chang, Y.L.; Wu, C.T.; Lee, Y.C.; Yang, S.C.; et al. Long form collapsin response mediator protein-1 (LCRMP-1) expression is associated with clinical outcome and lymph node metastasis in non-small cell lung cancer patients. Lung Cancer 2010, 67, 93–100. [Google Scholar] [CrossRef]
- Ji, X.; Woodard, A.S.; Rimm, D.L.; Fearon, E.R. Transcriptional defects underlie loss of E-cadherin expression in breast cancer. Cell Growth Differ. 1997, 8, 773–778. [Google Scholar] [PubMed]





| Variable | HR (95% CI) | p |
|---|---|---|
| Age | 1.00 (0.94–1.07) | 0.93 |
| Gender (male as reference) | 0.91 (0.26–3.22) | 0.88 |
| Smoking | 1.02 (0.27–3.80) | 0.98 |
| EGFR | 0.45 (0.15–1.31) | 0.14 |
| KRAS | 028 (0.03–2.29) | 0.23 |
| Id4 expression (per 1000 +) | 0.61 (0.41–0.90) | 0.012 |
| Slug expression (per 1000 +) | 1.32 (0.86–2.02) | 0.20 |
| E-cad expression (per 10 +) | 0.97 (0.85–1.12) | 0.69 |
| Variable | HR (95% CI) | p |
|---|---|---|
| Age | 1.01 (0.96–1.05) | 0.83 |
| Gender (male as reference) | 1.06 (0.43–2.60) | 0.89 |
| Smoking | 0.82 (0.32–2.07) | 0.67 |
| EGFR | 0.49 (0.23–1.06) | 0.069 |
| KRAS | 0.53 (0.17–1.68) | 0.28 |
| Id4 expression (per 1000 +) | 0.68 (0.54–0.86) | 0.001 |
| Slug expression (per 1000 +) | 1.73 (1.25–2.38) | 0.001 |
| E-cad expression (per 10 +) | 1.08 (1.01–1.16) | 0.035 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.-C.; Hsu, Y.-L.; Chang, C.-J.; Wang, C.-J.; Hsiao, T.-H.; Pan, S.-H. Inhibitor of DNA-Binding Protein 4 Suppresses Cancer Metastasis through the Regulation of Epithelial Mesenchymal Transition in Lung Adenocarcinoma. Cancers 2019, 11, 2021. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11122021
Wang C-C, Hsu Y-L, Chang C-J, Wang C-J, Hsiao T-H, Pan S-H. Inhibitor of DNA-Binding Protein 4 Suppresses Cancer Metastasis through the Regulation of Epithelial Mesenchymal Transition in Lung Adenocarcinoma. Cancers. 2019; 11(12):2021. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11122021
Chicago/Turabian StyleWang, Chi-Chung, Yuan-Ling Hsu, Chi-Jen Chang, Chia-Jen Wang, Tzu-Hung Hsiao, and Szu-Hua Pan. 2019. "Inhibitor of DNA-Binding Protein 4 Suppresses Cancer Metastasis through the Regulation of Epithelial Mesenchymal Transition in Lung Adenocarcinoma" Cancers 11, no. 12: 2021. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11122021