In Vitro Activity of 3-Bromopyruvate, an Anticancer Compound, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains
Abstract
:1. Introduction
2. Results
2.1. Disk-Diffusion Method
2.2. Determination of MICs and MBCs
2.3. Time-Killing Assay
2.4. Checkerboard Assay
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Disk-Diffusion Method
4.3. MIC/MBC Determination
4.4. Checkerboard Assay
4.5. Time-Killing Assay
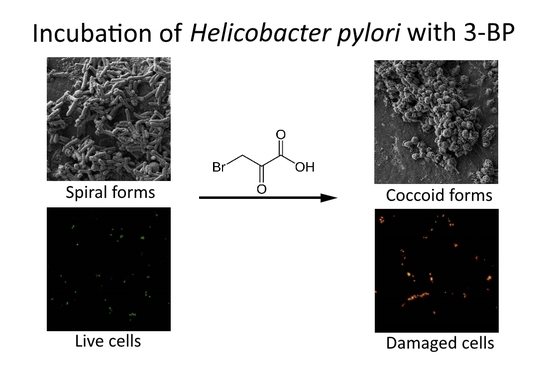
4.6. Light Microscopy
4.7. Fluorescence Microscopy
4.8. Scanning Electron Microscopy
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Díaz, P.; Valenzuela Valderrama, M.; Bravo, J.; Quest, A.F.G. Helicobacter pylori and Gastric Cancer: Adaptive Cellular Mechanisms Involved in Disease Progression. Front. Microbiol. 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P.; Kuipers, E.J.; Axon, A.T.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.Y.; et al. Management of Helicobacter pylori infection—The Maastricht V/Florence consensus report. Gut 2017, 66, 6–30. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Graham, D.Y. Helicobacter pylori Virulence and Cancer Pathogenesis. Futur. Oncol. 2014, 10, 1487–1500. [Google Scholar] [CrossRef] [PubMed]
- Watari, J.; Chen, N.; Amenta, P.S.; Fukui, H.; Oshima, T.; Tomita, T.; Miwa, H.; Lim, K.-J.; Das, K.M. Helicobacter pylori Associated Chronic Gastritis, Clinical Syndromes, Precancerous Lesions, and Pathogenesis of Gastric Cancer Development. World J. Gastroenterol. 2014, 20, 5461. [Google Scholar] [CrossRef] [PubMed]
- IARC Monographs on the Evaluation of Carcinogenic Risks to Humans Schistosomes, Liver Flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr. Eval. Carcinog. Risks Hum. 1994, 61, 1–241.
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global Cancer Statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Nagini, S. Carcinoma of The Stomach: A Review of Epidemiology, Pathogenesis, Molecular Genetics and Chemoprevention. World J. Gastrointest. Oncol. 2012, 4, 156–169. [Google Scholar] [CrossRef]
- Choi, J.M.; Kim, S.G.; Choi, J.; Park, J.Y.; Oh, S.; Yang, H.-J.; Lim, J.H.; Im, J.P.; Kim, J.S.; Jung, H.C. Effects of Helicobacter pylori Eradication for Metachronous Gastric Cancer Prevention: A Randomized Controlled Trial. Gastrointest. Endosc. 2018, 88, 475–485.e2. [Google Scholar] [CrossRef]
- Han, S.J.; Kim, S.G.; Lim, J.H.; Choi, J.M.; Oh, S.; Park, J.Y.; Kim, J.; Kim, J.S.; Jung, H.C. Long-Term Effects of Helicobacter pylori Eradication on Metachronous Gastric Cancer Development. Gut Liver 2018, 12, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.H.; Kim, J.-H.; Chung, H.S.; Park, J.C.; Shin, S.K.; Lee, S.K.; Lee, Y.C. Helicobacter pylori Eradication on the Prevention of Metachronous Lesions after Endoscopic Resection of Gastric Neoplasm: A Meta-Analysis. PLoS ONE 2015, 10, e0124725. [Google Scholar] [CrossRef] [PubMed]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Lis, P.; Dyląg, M.; Niedźwiecka, K.; Ko, Y.; Pedersen, P.; Goffeau, A.; Ułaszewski, S. The HK2 Dependent “Warburg Effect” and Mitochondrial Oxidative Phosphorylation in Cancer: Targets for Effective Therapy with 3-Bromopyruvate. Molecules 2016, 21, 1730. [Google Scholar] [CrossRef]
- Azevedo-Silva, J.; Queirós, O.; Baltazar, F.; Ułaszewski, S.; Goffeau, A.; Ko, Y.H.; Pedersen, P.L.; Preto, A.; Casal, M. The Anticancer Agent 3-Bromopyruvate: A Simple but Powerful Molecule Taken from the Lab to the Bedside. J. Bioenerg. Biomembr. 2016, 48, 349–362. [Google Scholar] [CrossRef]
- Ko, Y.H.; Pedersen, P.L.; Geschwind, J.F. Glucose Catabolism in the Rabbit VX2 Tumor Model for Liver Cancer: Characterization and Targeting Hexokinase. Cancer Lett. 2001, 173, 83–91. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, X.; Wang, T.; Xian, S.; Lu, Y. 3-Bromopyruvate and Sodium Citrate Induce Apoptosis in Human Gastric Cancer Cell Line MGC-803 by Inhibiting Glycolysis and Promoting Mitochondria-Regulated Apoptosis Pathway. Biochem. Biophys. Res. Commun. 2016, 475, 37–43. [Google Scholar] [CrossRef]
- Xian, S.-L.; Cao, W.; Zhang, X.-D.; Lu, Y.-F. Inhibitory Effects of 3-Bromopyruvate on Human Gastric Cancer Implant Tumors in Nude Mice. Asian Pacific J. Cancer Prev. 2014, 15, 3175–3178. [Google Scholar] [CrossRef] [Green Version]
- Barnard, J.P.; Reynafarje, B.; Pedersen, P.L. Glucose Catabolism in African Trypanosomes. Evidence that the Terminal Step is Catalyzed by a Pyruvate Transporter Capable of Facilitating Uptake of Toxic Analogs. J. Biol. Chem. 1993, 268, 3654–3661. [Google Scholar]
- de Lima, L.P.O.; Seabra, S.H.; Carneiro, H.; Barbosa, H.S. Effect of 3-Bromopyruvate and Atovaquone on Infection during In Vitro Interaction of Toxoplasma gondii and LLC-MK2 Cells. Antimicrob. Agents Chemother. 2015, 59, 5239–5249. [Google Scholar] [CrossRef] [PubMed]
- Lis, P.; Zarzycki, M.; Ko, Y.H.; Casal, M.; Pedersen, P.L.; Goffeau, A.; Ułaszewski, S. Transport and Cytotoxicity of the Anticancer Drug 3-Bromopyruvate in the Yeast Saccharomyces cerevisiae. J. Bioenerg. Biomembr. 2012, 44, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Dyląg, M.; Lis, P.; Niedźwiecka, K.; Ko, Y.H.; Pedersen, P.L.; Goffeau, A.; Ułaszewski, S. 3-Bromopyruvate: A Novel Antifungal Agent against the Human Pathogen Cryptococcus neoformans. Biochem. Biophys. Res. Commun. 2013, 434, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Jagielski, T.; Niedźwiecka, K.; Roeske, K.; Dyląg, M. 3-Bromopyruvate as an Alternative Option for the Treatment of Protothecosis. Front. Pharmacol. 2018, 9, 375. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.S.; Singh, S.K. The Mycobacterium tuberculosis H37Ra Gene MRA_1916 Causes Growth Defects upon Down-Regulation. Sci. Rep. 2015, 5, 16131. [Google Scholar] [CrossRef]
- Visca, P.; Pisa, F.; Imperi, F. The Antimetabolite 3-Bromopyruvate Selectively Inhibits Staphylococcus aureus. Int. J. Antimicrob. Agents 2018. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Yamaoka, Y. Drug Resistance in Helicobacter pylori. Adv. Res. Gastroenterol. Hepatol. 2017, 7. ARGH.MS.ID.555715. [Google Scholar]
- Ghotaslou, R.; Leylabadlo, H.E.; Asl, Y.M. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Recent Literature Review. World J. Methodol. 2015, 5, 164–174. [Google Scholar] [CrossRef]
- Megraud, F.; Coenen, S.; Versporten, A.; Kist, M.; Lopez-Brea, M.; Hirschl, A.M.; Andersen, L.P.; Goossens, H.; Glupczynski, Y.; Study Group participants. Helicobacter pylori Resistance to Antibiotics in Europe and Its Relationship to Antibiotic Consumption. Gut 2013, 62, 34–42. [Google Scholar] [CrossRef]
- Boltin, D.; Ben-Zvi, H.; Perets, T.T.; Kamenetsky, Z.; Samra, Z.; Dickman, R.; Niv, Y. Trends in Secondary Antibiotic Resistance of Helicobacter pylori from 2007 to 2014: Has the Tide Turned? J. Clin. Microbiol. 2015, 53, 522–527. [Google Scholar] [CrossRef]
- Bisignano, C.; Filocamo, A.; La Camera, E.; Zummo, S.; Fera, M.T.; Mandalari, G. Antibacterial Activities of Almond Skins on cagA-Positive and-Negative Clinical Isolates of Helicobacter pylori. BMC Microbiol. 2013, 13, 103. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Hsu, C.; Yin, M. In Vitro Anti-Helicobacter pylori Activity of Diallyl Sulphides and Protocatechuic Acid. Phyther. Res. 2008, 22, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Obonyo, M.; Zhang, L.; Thamphiwatana, S.; Pornpattananangkul, D.; Fu, V.; Zhang, L. Antibacterial Activities of Liposomal Linolenic Acids against Antibiotic-Resistant Helicobacter pylori. Mol. Pharm. 2012, 9, 2677–2685. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Haristoy, X.; Dolan, P.M.; Kensler, T.W.; Scholtus, I.; Stephenson, K.K.; Talalay, P.; Lozniewski, A. Sulforaphane Inhibits Extracellular, Intracellular, and Antibiotic-Resistant Strains of Helicobacter pylori and Prevents Benzo[a]pyrene-Induced Stomach Tumors. Proc. Natl. Acad. Sci. USA 2002, 99, 7610–7615. [Google Scholar] [CrossRef] [PubMed]
- Vali, M.; Liapi, E.; Kowalski, J.; Hong, K.; Khwaja, A.; Torbenson, M.S.; Georgiades, C.; Geschwind, J.-F.H. Intraarterial Therapy with a New Potent Inhibitor of Tumor Metabolism (3-Bromopyruvate): Identification of Therapeutic Dose and Method of Injection in an Animal Model of Liver Cancer. J. Vasc. Interv. Radiol. 2007, 18, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Kunjithapatham, R.; Geschwind, J.-F.H.; Rao, P.P.; Boronina, T.N.; Cole, R.N.; Ganapathy-Kanniappan, S. Systemic Administration of 3-Bromopyruvate Reveals Its Interaction with Serum Proteins in a Rat Model. BMC Res. Notes 2013, 6, 277. [Google Scholar] [CrossRef] [PubMed]
- Levison, M.E. Pharmacodynamics of Antimicrobial Drugs. Infect. Dis. Clin. North Am. 2004, 18, 451–465. [Google Scholar] [CrossRef]
- Krzyżek, P.; Gościniak, G. Morphology of Helicobacter pylori as a Result of Peptidoglycan and Cytoskeleton Rearrangements. Prz. Gastroenterol. 2018, 13, 182–195. [Google Scholar] [CrossRef]
- Sarem, M.; Corti, R. Role of Helicobacter pylori Coccoid Forms in Infection and Recrudescence. Gastroenterol. Hepatol. 2016, 39, 28–35. [Google Scholar] [CrossRef]
- Cellini, L. Helicobacter pylori: A Chameleon-Like Approach to Life. World J. Gastroenterol. 2014, 20, 5575–5582. [Google Scholar] [CrossRef]
- Krzyżek, P.; Biernat, M.M.; Gościniak, G. Intensive Formation of Coccoid Forms as a Feature Strongly Associated with Highly Pathogenic Helicobacter pylori Strains. Folia Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-K.; Song, H.E.; Lee, H.-B.; Kim, C.-S.; Koketsu, M.; Thi My Ngan, L.; Ahn, Y.-J. Growth Inhibitory, Bactericidal, and Morphostructural Effects of Dehydrocostus Lactone from Magnolia sieboldii Leaves on Antibiotic-Susceptible and -Resistant Strains of Helicobacter pylori. PLoS ONE 2014, 9, e95530. [Google Scholar] [CrossRef] [PubMed]
- Ngan, L.T.M.; Moon, J.-K.; Shibamoto, T.; Ahn, Y.-J. Growth-Inhibiting, Bactericidal, and Urease Inhibitory Effects of Paeonia lactiflora Root Constituents and Related Compounds on Antibiotic-Susceptible and -Resistant Strains of Helicobacter pylori. J. Agric. Food Chem. 2012, 60, 9062–9073. [Google Scholar] [CrossRef] [PubMed]
- Faghri, J.; Poursina, F.; Moghim, S.; Zarkesh Esfahani, H.; Nasr Esfahani, B.; Fazeli, H.; Mirzaei, N.; Jamshidian, A.; Ghasemian Safaei, H. Morphological and Bactericidal Effects of Different Antibiotics on Helicobacter pylori. Jundishapur J. Microbiol. 2014, 7, e8704. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The Importance of the Viable but Non-Culturable State in Human Bacterial Pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.-S.; Ou, K.-L.; Peng, P.-W.; Liou, B.-C.; Wang, W.-T.; Huang, Y.-C.; Tsai, C.-M.; Su, C.-H. Quantifying Membrane Permeability of Amphotericin B Ion Channels in Single Living Cells. Biochim. Biophys. Acta 2013, 1828, 1794–1801. [Google Scholar] [CrossRef] [PubMed]
- Novo, D.J.; Perlmutter, N.G.; Hunt, R.H.; Shapiro, H.M. Multiparameter Flow Cytometric Analysis of Antibiotic Effects on Membrane Potential, Membrane Permeability, and Bacterial Counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob. Agents Chemother. 2000, 44, 827–834. [Google Scholar] [CrossRef] [Green Version]
- Novickij, V.; Švedienė, J.; Paškevičius, A.; Markovskaja, S.; Lastauskienė, E.; Zinkevičienė, A.; Girkontaitė, I.; Novickij, J. Induction of Different Sensitization Patterns of MRSA to Antibiotics Using Electroporation. Molecules 2018, 23, 1799. [Google Scholar] [CrossRef]
- Tateda, K.; Ishii, Y.; Matsumoto, T.; Kobayashi, T.; Miyazaki, S.; Yamaguchi, K. Potential of Macrolide Antibiotics to Inhibit Protein Synthesis of Pseudomonas aeruginosa: Suppression of Virulence Factors and Stress Response. J. Infect. Chemother. 2000, 6, 1–7. [Google Scholar] [CrossRef]
- Gościniak, G.; Biernat, M.; Grabińska, J.; Bińkowska, A.; Poniewierka, E.; Iwańczak, B. The Antimicrobial Susceptibility of Helicobacter pylori Strains Isolated from Children and Adults with Primary Infection in the Lower Silesia Region, Poland. Pol. J. Microbiol. 2014, 63, 57–61. [Google Scholar]
- Gościniak, G.; Biernat, M.M.; Bińkowska, A.; Kus, A.; Iwańczak, B. Frequency of Infection with Helicobacter pylori Isolates of Different Antimicrobial Profiles in Children and Adolescents: A Preliminary Study. Adv. Clin. Exp. Med. 2017, 26, 263–268. [Google Scholar] [PubMed]
- EUCAST (The European Committee on Antimicrobial Susceptibility Testing). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 5.0; EUCAST: Växjö, Sweden, 2015. [Google Scholar]
- Chisholm, S.A.; Owen, R.J. Frequency and Molecular Characteristics of Ciprofloxacin- and Rifampicin-resistant Helicobacter pylori from Gastric Infections in the UK. J. Med. Microbiol. 2009, 58, 1322–1328. [Google Scholar] [CrossRef]
- Boyanova, L.; Gergova, G.; Nikolov, R.; Derejian, S.; Lazarova, E.; Katsarov, N.; Mitov, I.; Krastev, Z. Activity of Bulgarian Propolis against 94 Helicobacter pylori strains In Vitro by Agar-Well Diffusion, Agar Dilution and Disc Diffusion Methods. J. Med. Microbiol. 2005, 54, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, H.; Osaki, T.; Hanawa, T.; Kurata, S.; Ochiai, K.; Kamiya, S. Impact of Helicobacter pylori Biofilm Formation on Clarithromycin Susceptibility and Generation of Resistance Mutations. PLoS ONE 2013, 8, e73301. [Google Scholar] [CrossRef]
- Cellini, L.; Di Bartolomeo, S.; Di Campli, E.; Genovese, S.; Locatelli, M.; Di Giulio, M. In vitro activity of Aloe vera inner gel against Helicobacter pylori strains. Lett. Appl. Microbiol. 2014, 59, 43–48. [Google Scholar] [CrossRef]
- Hirschl, A.M.; Apfalter, P.; Makristathis, A.; Rotter, M.L.; Wimmer, M. In Vitro Activities of Linezolid Alone and in Combination with Amoxicillin, Clarithromycin, and Metronidazole against Helicobacter pylori. Antimicrob. Agents Chemother. 2000, 44, 1977–1979. [Google Scholar] [CrossRef]
- Nostro, A.; Cellini, L.; Di Bartolomeo, S.; Cannatelli, M.A.; Di Campli, E.; Procopio, F.; Grande, R.; Marzio, L.; Alonzo, V. Effects of Combining Extracts (from Propolis or Zingiber officinale) with Clarithromycin on Helicobacter pylori. Phyther. Res. 2006, 20, 187–190. [Google Scholar] [CrossRef]
- Abu-Qatouseh, L.; Abu-Sini, M.; Mayyas, A.; Al-Hiari, Y.; Darwish, R.; Aburjai, T. Synthesis of New Nitrofluoroquinolone Derivatives with Novel Anti-Microbial Properties against Metronidazole Resistant H. pylori. Molecules 2017, 22, 71. [Google Scholar] [CrossRef]
- Brown, J.C.; Jiang, X. Activities of Muscadine Grape Skin and Polyphenolic Constituents against Helicobacter pylori. J. Appl. Microbiol. 2013, 114, 982–991. [Google Scholar] [CrossRef]
- Chou, T.-M.; Ke, Y.-Y.; Tsao, Y.-H.; Li, Y.-C.; Lin, Z.-H. Fabrication of Te and Te-Au Nanowires-Based Carbon Fiber Fabrics for Antibacterial Applications. Int. J. Environ. Res. Public Health 2016, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Marchesini, M.I.; Morrone Seijo, S.M.; Guaimas, F.F.; Comerci, D.J. A T4SS Effector Targets Host Cell Alpha-Enolase Contributing to Brucella abortus Intracellular Lifestyle. Front. Cell. Infect. Microbiol. 2016, 6, 153. [Google Scholar] [CrossRef] [PubMed]












| Strains | Antibiotic Resistance * | 3-BP | AMX ** | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 200 µg/disk | 1000 µg/disk | 2000 µg/disk | 25 µg/disk | |||||||||||
| MTZ | CLR | Sample | Average | Sample | Average | Sample | Average | Sample | Average | |||||
| I | II | I | II | I | II | I | II | |||||||
| 1950 | R | R | 6 | 6 | 6 | 17 | 21 | 19 | 21 | 24 | 22,5 | 69 | 70 | 69.5 |
| 1952 | S | R | 11 | 13 | 12 | 20 | 23 | 21.5 | 23 | 21 | 22 | 67 | 71 | 69 |
| 1954 | R | R | 13 | 12 | 12.5 | 13 | 13 | 13 | 18 | 22 | 20 | 68 | 67 | 67.5 |
| 1964 | R | R | 6 | 6 | 6 | 14 | 15 | 14.5 | 24 | 20 | 22 | 70 | 71 | 70.5 |
| 2093 | R | S | 8 | 10 | 9 | 11 | 12 | 11.5 | 16 | 16 | 16 | 62 | 64 | 63 |
| 2095 | S | S | 11 | 10 | 10.5 | 18 | 15 | 16.5 | 25 | 28 | 26.5 | 70 | 69 | 69.5 |
| 6010 | R | S | 9 | 10 | 9.5 | 14 | 15 | 14.5 | 18 | 21 | 19.5 | 66 | 65 | 65.5 |
| 6171 | S | S | 10 | 10 | 10 | 14 | 14 | 14 | 18 | 17 | 17.5 | 65 | 65 | 65 |
| 6237 | S | S | 9 | 10 | 9.5 | 18 | 17 | 17.5 | 26 | 28 | 27 | 68 | 72 | 70 |
| 6291 | R | R | 16 | 15 | 15.5 | 23 | 25 | 24 | 26 | 27 | 26.5 | 69 | 70 | 69.5 |
| 6341 | R | R | 9 | 9 | 9 | 25 | 21 | 23 | 34 | 31 | 32.5 | 71 | 70 | 70.5 |
| 6343 | S | S | 7 | 9 | 8 | 19 | 21 | 20 | 29 | 32 | 30.5 | 69 | 71 | 70 |
| 6522 | R | R | 7 | 8 | 7.5 | 15 | 16 | 15.5 | 20 | 18 | 19 | 65 | 64 | 64.5 |
| 6559 | R | S | 8 | 8 | 8 | 17 | 16 | 16.5 | 28 | 31 | 29.5 | 66 | 69 | 67.5 |
| 6574 | S | S | 8 | 8 | 8 | 14 | 16 | 15 | 17 | 19 | 18 | 60 | 64 | 62 |
| 6575 | S | S | 8 | 9 | 8.5 | 15 | 15 | 15 | 29 | 33 | 31 | 64 | 63 | 63.5 |
| 6638 | S | R | 8 | 8 | 8 | 16 | 15 | 15.5 | 27 | 30 | 28.5 | 70 | 69 | 69.5 |
| 6649 | S | S | 6 | 7 | 6.5 | 13 | 14 | 13.5 | 18 | 18 | 18 | 66 | 66 | 66 |
| 6653 | S | R | 7 | 7 | 7 | 13 | 13 | 13 | 25 | 29 | 27 | 68 | 70 | 69 |
| 6687 | R | R | 7 | 8 | 7.5 | 15 | 16 | 15.5 | 20 | 23 | 21.5 | 68 | 69 | 68.5 |
| 6699 | S | R | 7 | 9 | 8 | 18 | 20 | 19 | 22 | 25 | 23.5 | 69 | 70 | 69.5 |
| 6716 | S | S | 18 | 20 | 19 | 29 | 28 | 28.5 | 30 | 32 | 31 | 71 | 70 | 70.5 |
| 6735 | S | S | 10 | 10 | 10 | 15 | 15 | 15 | 22 | 24 | 23 | 67 | 68 | 67.5 |
| 6741 | S | S | 9 | 10 | 9.5 | 13 | 14 | 13.5 | 19 | 19 | 19 | 63 | 65 | 64 |
| 6794 | R | R | 9 | 8 | 8.5 | 12 | 12 | 12 | 17 | 19 | 18 | 63 | 66 | 64.5 |
| 6858 | S | R | 7 | 7 | 7 | 13 | 14 | 13.5 | 17 | 17 | 17 | 61 | 58 | 59.5 |
| 6875 | R | S | 9 | 10 | 9.5 | 12 | 12 | 12 | 18 | 19 | 18.5 | 68 | 72 | 70 |
| 6885 | S | S | 6 | 6 | 6 | 10 | 10 | 10 | 16 | 17 | 16.5 | 69 | 72 | 70.5 |
| 7042 | S | S | 11 | 9 | 10 | 13 | 14 | 13.5 | 18 | 17 | 17.5 | 61 | 64 | 62.5 |
| 7080 | R | S | 10 | 12 | 11 | 15 | 13 | 14 | 16 | 17 | 16.5 | 62 | 62 | 62 |
| 7101 | S | S | 9 | 10 | 9.5 | 13 | 14 | 13.5 | 18 | 18 | 18 | 65 | 64 | 64.5 |
| 7110 | S | S | 11 | 11 | 11 | 14 | 16 | 15 | 22 | 23 | 22.5 | 67 | 68 | 67.5 |
| 7143 | R | R | 8 | 8 | 8 | 13 | 13 | 13 | 18 | 17 | 17.5 | 66 | 67 | 66.5 |
| 7173 | R | S | 9 | 11 | 10 | 13 | 14 | 13.5 | 17 | 17 | 17 | 60 | 62 | 61 |
| 7189 | S | R | 11 | 10 | 10.5 | 12 | 12 | 12 | 20 | 18 | 19 | 64 | 63 | 63.5 |
| 7208 | S | S | 8 | 9 | 8.5 | 14 | 15 | 14.5 | 21 | 24 | 22.5 | 67 | 63 | 65 |
| 7264 | S | S | 9 | 9 | 9 | 11 | 13 | 12 | 17 | 18 | 17.5 | 69 | 70 | 69.5 |
| 7286 | R | S | 8 | 9 | 8.5 | 14 | 14 | 14 | 19 | 17 | 18 | 69 | 68 | 68.5 |
| 7297 | S | S | 10 | 11 | 10.5 | 17 | 17 | 17 | 20 | 22 | 21 | 63 | 61 | 62 |
| 7308 | S | S | 9 | 9 | 9 | 12 | 12 | 12 | 18 | 19 | 18.5 | 57 | 59 | 58 |
| 7317 | S | R | 11 | 12 | 11.5 | 15 | 14 | 14.5 | 19 | 19 | 19 | 59 | 60 | 59.5 |
| 7357 | R | S | 11 | 10 | 10.5 | 16 | 17 | 16.5 | 19 | 20 | 19.5 | 69 | 70 | 69.5 |
| 7361 | R | S | 9 | 9 | 9 | 15 | 15 | 15 | 19 | 18 | 18.5 | 69 | 71 | 70 |
| 7388 | R | S | 8 | 8 | 8 | 13 | 14 | 13.5 | 17 | 17 | 17 | 71 | 70 | 70.5 |
| 7394 | R | S | 14 | 13 | 13.5 | 26 | 24 | 25 | 29 | 33 | 31 | 67 | 64 | 65.5 |
| 7404 | S | S | 11 | 10 | 10.5 | 17 | 17 | 17 | 24 | 27 | 25.5 | 68 | 67 | 67.5 |
| 7471 | S | S | 12 | 13 | 12.5 | 21 | 22 | 21.5 | 27 | 30 | 28.5 | 57 | 60 | 58.5 |
| 7556 | S | R | 17 | 15 | 16 | 24 | 23 | 23.5 | 29 | 27 | 28 | 71 | 70 | 70.5 |
| 7649 | R | R | 10 | 10 | 10 | 16 | 16 | 16 | 25 | 26 | 25.5 | 64 | 61 | 62.5 |
| 8064 | R | R | 7 | 8 | 7.5 | 16 | 15 | 15.5 | 20 | 22 | 21 | 60 | 62 | 61 |
| J99 | S | S | 6 | 6 | 6 | 11 | 11 | 11 | 19 | 18 | 18.5 | 65 | 68 | 66.5 |
| Tx30a | S | S | 10 | 12 | 11 | 14 | 15 | 14.5 | 24 | 23 | 23.5 | 68 | 71 | 69.5 |
| Strains | Antibiotic Resistance * | Activity of 3-BP | |||
|---|---|---|---|---|---|
| MTZ | CLR | MIC** | MBC ** | MBC/MIC Ratio | |
| J99 | S | S | 128 | 128 | 1 |
| Tx30a | S | S | 128 | 128 | 1 |
| 6237 | S | S | 32 | 128 | 4 |
| 7471 | S | S | 64 | 128 | 2 |
| 7189 | S | R | 32 | 128 | 4 |
| 7556 | S | R | 128 | 128 | 1 |
| 7388 | R | S | 128 | 128 | 1 |
| 7394 | R | S | 64 | 128 | 2 |
| 7143 | R | R | 128 | 128 | 1 |
| 7649 | R | R | 128 | 128 | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krzyżek, P.; Franiczek, R.; Krzyżanowska, B.; Łaczmański, Ł.; Migdał, P.; Gościniak, G. In Vitro Activity of 3-Bromopyruvate, an Anticancer Compound, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains. Cancers 2019, 11, 229. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11020229
Krzyżek P, Franiczek R, Krzyżanowska B, Łaczmański Ł, Migdał P, Gościniak G. In Vitro Activity of 3-Bromopyruvate, an Anticancer Compound, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains. Cancers. 2019; 11(2):229. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11020229
Chicago/Turabian StyleKrzyżek, Paweł, Roman Franiczek, Barbara Krzyżanowska, Łukasz Łaczmański, Paweł Migdał, and Grażyna Gościniak. 2019. "In Vitro Activity of 3-Bromopyruvate, an Anticancer Compound, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains" Cancers 11, no. 2: 229. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11020229