SNAIL Promotes Metastatic Behavior of Rhabdomyosarcoma by Increasing EZRIN and AKT Expression and Regulating MicroRNA Networks
Abstract
:1. Introduction
2. Results
2.1. SNAIL Regulates the Metastatic Behavior of RMS Cells In Vivo and In Vitro
2.2. EZRIN is a Crucial Mediator of the Metastatic Action of SNAIL in RMS Cells
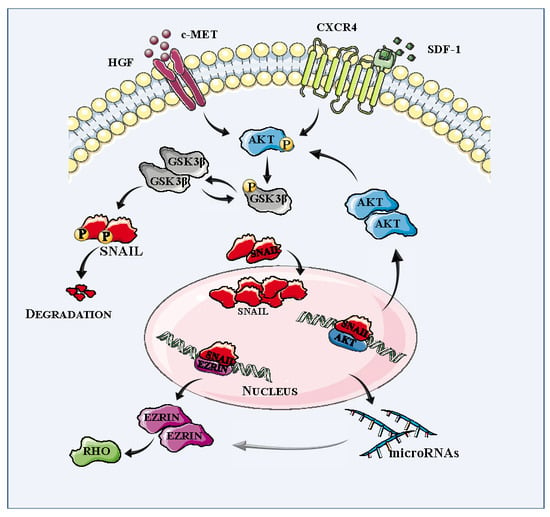
2.3. Mutual Regulation of SNAIL and AKT Kinase Expression in RMS Cells
2.4. Inhibition of PI3K-AKT Signaling Diminishes Migration and Chemotaxis of RMS Cells
2.5. The SNAIL-miRNA Axis Regulates the Motility of RMS Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Treatments of the Cells
4.3. Production of Viral Vectors and Transduction of Cells
4.4. Transfection with siRNA
4.5. Transfection of Cells with miRNA Precursors and Inhibitors
4.6. DNA and RNA Isolation and Reverse Transcription
4.7. Quantitative Real-Time PCR
4.8. MicroRNA Sequencing
4.9. Western Blotting
4.10. RHO and ROCK-II Enzymes Activity
4.11. Analysis of Subproteomes of Nuclear and Cytoplasmic Fractions
4.12. Bioinformatic Analysis
4.13. Chromatin Immunoprecipitation (ChIP) Assay
- EZR_1 fragment forward primer: 5′-GAGGCTAGCACGAGTTAAGCA-3′
- EZR_1 fragment reverse primer: 5′-GCACGTTTGTGGCCTCTTTT-3′
- EZR_2 fragment forward primer: 5′-GGAGCACACGGAGCACTG-3′
- EZR_2 fragment reverse primer: 5′-CGGAGAGAGGCGGAGAAGA-3′
4.14. Scratch Assay
4.15. Chemotaxis and Invasion Assays
4.16. Immunofluorescent Staining with Phalloidin
4.17. Flow Cytometry
4.18. Adhesion Assay
4.19. In Vivo Experiments
4.20. Bioinformatical Analysis of Microarray Data from RMS Patients
4.21. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Prim. 2019, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K.A.; Wexler, L.H. Pediatric rhabdomyosarcoma with bone marrow metastasis. Pediatr. Blood Cancer 2020. [Google Scholar] [CrossRef]
- Jankowski, K.; Kucia, M.; Wysoczynski, M.; Reca, R.; Zhao, D.; Trzyna, E.; Trent, J.; Peiper, S.; Zembala, M.; Ratajczak, J.; et al. Both Hepatocyte Growth Factor (HGF) and Stromal-Derived Factor-1 Regulate the Metastatic Behavior of Human Rhabdomyosarcoma Cells, but only HGF Enhances Their Resistance to Radiochemotherapy. Cancer Res. 2003, 63, 7926–7935. [Google Scholar]
- Libura, J.; Drukala, J.; Majka, M.; Tomescu, O.; Navenot, J.M.; Kucia, M.; Marquez, L.; Peiper, S.C.; Barr, F.G.; Janowska-Wieczorek, A.; et al. CXCR4-SDF-1 signaling is active in rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and adhesion. Blood 2002, 100, 2597–2606. [Google Scholar] [CrossRef] [Green Version]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Hettmer, S.; Wagers, A.J. Muscling in: Uncovering the origins of rhabdomyosarcoma. Nat. Med. 2010, 16, 171–173. [Google Scholar] [CrossRef]
- Szewczyk, B.; Skrzypek, K.; Majka, M. Targeting MET Receptor in Rhabdomyosarcoma: Rationale and Progress. Curr. Drug Targets 2017, 18, 98–107. [Google Scholar] [CrossRef]
- Yu, Y.; Khan, J.; Khanna, C.; Helman, L.; Meltzer, P.S.; Merlino, G. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat. Med. 2004, 10, 175–181. [Google Scholar] [CrossRef]
- Skrzypek, K.; Kusienicka, A.; Trzyna, E.; Szewczyk, B.; Ulman, A.; Konieczny, P.; Adamus, T.; Badyra, B.; Kortylewski, M.; Majka, M. SNAIL is a key regulator of alveolar rhabdomyosarcoma tumor growth and differentiation through repression of MYF5 and MYOD function. Cell Death Dis. 2018, 9, 643. [Google Scholar] [CrossRef] [PubMed]
- Ulman, A.; Skrzypek, K.; Konieczny, P.; Mussolino, C.; Cathomen, T.; Majka, M. Genome Editing of the SNAI1 Gene in Rhabdomyosarcoma: A Novel Model for Studies of Its Role. Cells 2020, 9, 1095. [Google Scholar] [CrossRef]
- Püsküllüoglu, M.; Lukasiewicz, E.; Miekus, K.; Jarocha, D.; Majka, M. Differential expression of Snail1 transcription factor and Snail1-related genes in alveolar and embryonal rhabdomyosarcoma subtypes. Folia Histochem. Cytobiol. 2010, 48, 671–677. [Google Scholar] [PubMed] [Green Version]
- Xu, L.; Zheng, Y.; Liu, J.; Rakheja, D.; Singleterry, S.; Laetsch, T.W.; Shern, J.F.; Khan, J.; Triche, T.J.; Hawkins, D.S.; et al. Integrative Bayesian Analysis Identifies Rhabdomyosarcoma Disease Genes. Cell Rep. 2018, 24, 238–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peinado, H.; Ballestar, E.; Esteller, M.; Cano, A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell Biol. 2004, 24, 306–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef]
- Nieszporek, A.; Skrzypek, K.; Adamek, G.; Majka, M. Molecular mechanisms of epithelial to mesenchymal transition in tumor metastasis. Acta Biochim. Pol. 2019, 66, 509–520. [Google Scholar] [CrossRef] [Green Version]
- Barrallo-Gimeno, A.; Nieto, M.A. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development 2005, 132, 3151–3161. [Google Scholar] [CrossRef] [Green Version]
- Skrzypek, K.; Majka, M. Interplay among SNAIL Transcription Factor, MicroRNAs, Long Non-Coding RNAs, and Circular RNAs in the Regulation of Tumor Growth and Metastasis. Cancers 2020, 12, 209. [Google Scholar] [CrossRef] [Green Version]
- Olmeda, D.; Jordá, M.; Peinado, H.; Fabra, Á.; Cano, A. Snail silencing effectively suppresses tumour growth and invasiveness. Oncogene 2007, 26, 1862–1874. [Google Scholar] [CrossRef] [Green Version]
- Lambies, G.; Miceli, M.; Martínez-Guillamon, C.; Olivera-Salguero, R.; Peña, R.; Frías, C.-P.; Calderón, I.; Atanassov, B.S.; Dent, S.Y.R.; Arribas, J.; et al. TGFβ-Activated USP27X Deubiquitinase Regulates Cell Migration and Chemoresistance via Stabilization of Snail1. Cancer Res. 2019, 79, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Batlle, R.; Alba-Castellón, L.; Loubat-Casanovas, J.; Armenteros, E.; Francí, C.; Stanisavljevic, J.; Banderas, R.; Martin-Caballero, J.; Bonilla, F.; Baulida, J.; et al. Snail1 controls TGF-β responsiveness and differentiation of mesenchymal stem cells. Oncogene 2013, 32, 3381–3389. [Google Scholar] [CrossRef] [Green Version]
- Skrzypek, K.; Kusienicka, A.; Szewczyk, B.; Adamus, T.; Lukasiewicz, E.; Miekus, K.; Majka, M. Constitutive activation of MET signaling impairs myogenic differentiation of rhabdomyosarcoma and promotes its development and progression. Oncotarget 2015, 6, 31378–31398. [Google Scholar] [CrossRef] [Green Version]
- Alba-Castellón, L.; Batlle, R.; Francí, C.; Fernández-Aceñero, M.J.; Mazzolini, R.; Peña, R.; Loubat, J.; Alameda, F.; Rodríguez, R.; Curto, J.; et al. Snail1 expression is required for sarcomagenesis. Neoplasia 2014, 16, 413–421. [Google Scholar] [CrossRef] [Green Version]
- Ignatius, M.S.; Hayes, M.N.; Lobbardi, R.; Chen, E.Y.; McCarthy, K.M.; Sreenivas, P.; Motala, Z.; Durbin, A.D.; Molodtsov, A.; Reeder, S.; et al. The NOTCH1/SNAIL1/MEF2C Pathway Regulates Growth and Self-Renewal in Embryonal Rhabdomyosarcoma. Cell Rep. 2017, 19, 2304–2318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef]
- Shankar, J.; Nabi, I.R. Actin Cytoskeleton Regulation of Epithelial Mesenchymal Transition in Metastatic Cancer Cells. PLoS ONE 2015, 10, e0119954. [Google Scholar]
- Petty, J.M.; Sueblinvong, V.; Lenox, C.C.; Jones, C.C.; Cosgrove, G.P.; Cool, C.D.; Rai, P.R.; Brown, K.K.; Weiss, D.J.; Poynter, M.E.; et al. Pulmonary Stromal-Derived Factor-1 Expression and Effect on Neutrophil Recruitment during Acute Lung Injury. J. Immunol. 2007, 178, 8148–8157. [Google Scholar] [CrossRef] [PubMed]
- Svitkina, T. The actin cytoskeleton and actin-based motility. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Jiang, H.; Zhao, W.; Meng, Y.; Li, J.; Huang, T.; Sun, J. Cdc42-mediated supracellular cytoskeleton induced cancer cell migration under low shear stress. Biochem. Biophys. Res. Commun. 2019, 519, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Mattila, P.K.; Lappalainen, P. Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Dreos, R.; Ambrosini, G.; Groux, R.; Cavin Périer, R.; Bucher, P. The eukaryotic promoter database in its 30th year: Focus on non-vertebrate organisms. Nucleic Acids Res. 2017, 45, D51–D55. [Google Scholar] [CrossRef] [Green Version]
- Kohsaka, S.; Shukla, N.; Ameur, N.; Ito, T.; Ng, C.K.Y.; Wang, L.; Lim, D.; Marchetti, A.; Viale, A.; Pirun, M.; et al. A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat. Genet. 2014, 46, 595–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villagrasa, P.; Díaz, V.M.; Viñas-Castells, R.; Peiró, S.; Del Valle-Pérez, B.; Dave, N.; Rodríguez-Asiain, A.; Casal, J.I.; Lizcano, J.M.; Duñach, M.; et al. Akt2 interacts with Snail1 in the E-cadherin promoter. Oncogene 2012, 31, 4022–4033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frías, A.; Lambies, G.; Viñas-Castells, R.; Martínez-Guillamon, C.; Dave, N.; García de Herreros, A.; Díaz, V.M. A Switch in Akt Isoforms Is Required for Notch-Induced Snail1 Expression and Protection from Cell Death. Mol. Cell. Biol. 2016, 36, 923–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermida, M.A.; Dinesh Kumar, J.; Leslie, N.R. GSK3 and its interactions with the PI3K/AKT/mTOR signalling network. Adv. Biol. Regul. 2017, 65, 5–15. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.P.; Deng, J.; Xia, W.; Xu, J.; Li, Y.M.; Gunduz, M.; Hung, M.-C. Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial–mesenchymal transition. Nat. Cell Biol. 2004, 6, 931–940. [Google Scholar] [CrossRef]
- Ye, X.; Weinberg, R.A. The SUMO guards for SNAIL. Oncotarget 2017, 8, 97701–97702. [Google Scholar] [CrossRef]
- Peiro, S.; Escrivà, M.; Puig, I.; Barberà, M.J.; Dave, N.; Herranz, N.; Larriba, M.J.; Takkunen, M.; Francí, C.; Muñoz, A.; et al. Snail1 transcriptional repressor binds to its own promoter and controls its expression. Nucleic Acids Res. 2006, 34, 2077–2084. [Google Scholar] [CrossRef]
- Fröse, J.; Chen, M.B.; Hebron, K.E.; Reinhardt, F.; Hajal, C.; Zijlstra, A.; Kamm, R.D.; Weinberg, R.A. Epithelial-Mesenchymal Transition Induces Podocalyxin to Promote Extravasation via Ezrin Signaling. Cell Rep. 2018, 24, 962–972. [Google Scholar] [CrossRef] [Green Version]
- Curto, M.; McClatchey, A.I. Ezrin...a metastatic detERMinant? Cancer Cell 2004, 5, 113–114. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.; Hong, S.H.; Cassavaugh, J.; Osborne, T.; Chou, A.J.; Kim, S.Y.; Gorlick, R.; Hewitt, S.M.; Khanna, C. The actin-cytoskeleton linker protein ezrin is regulated during osteosarcoma metastasis by PKC. Oncogene 2009, 28, 792–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croft, D.R.; Sahai, E.; Mavria, G.; Li, S.; Tsai, J.; Lee, W.M.F.; Marshall, C.J.; Olson, M.F. Conditional ROCK Activation In vivo Induces Tumor Cell Dissemination and Angiogenesis. Cancer Res. 2004, 64, 8994–9001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahai, E.; Marshall, C.J. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 2003, 5, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, C.-X.; Gao, Y. Snail regulates the motility of oral cancer cells via RhoA/Cdc42/p-ERM pathway. Biochem. Biophys. Res. Commun. 2014, 452, 490–496. [Google Scholar] [CrossRef]
- Tao, G.; Levay, A.K.; Gridley, T.; Lincoln, J. Mmp15 is a direct target of Snai1 during endothelial to mesenchymal transformation and endocardial cushion development. Dev. Biol. 2011, 359, 209–221. [Google Scholar] [CrossRef] [Green Version]
- Rembold, M.; Ciglar, L.; Yáñez-Cuna, J.O.; Zinzen, R.P.; Girardot, C.; Jain, A.; Welte, M.A.; Stark, A.; Leptin, M.; Furlong, E.E.M. A conserved role for Snail as a potentiator of active transcription. Genes Dev. 2014, 28, 167. [Google Scholar] [CrossRef] [Green Version]
- Subhra Das, S.; James, M.; Paul, S.; Chakravorty, N. miRnalyze: An interactive database linking tool to unlock intuitive microRNA regulation of cell signaling pathways. Database (Oxford) 2017, 2017. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Bartolomé-Izquierdo, N.; de Yébenes, V.G.; Álvarez-Prado, A.F.; Mur, S.M.; Lopez del Olmo, J.A.; Roa, S.; Vazquez, J.; Ramiro, A.R. miR-28 regulates the germinal center reaction and blocks tumor growth in preclinical models of non-Hodgkin lymphoma. Blood 2017, 129, 2408–2419. [Google Scholar] [CrossRef]
- Almeida, M.I.; Nicoloso, M.S.; Zeng, L.; Ivan, C.; Spizzo, R.; Gafà, R.; Xiao, L.; Zhang, X.; Vannini, I.; Fanini, F.; et al. Strand-Specific miR-28-5p and miR-28-3p Have Distinct Effects in Colorectal Cancer Cells. Gastroenterology 2012, 142, 886–896.e9. [Google Scholar] [CrossRef] [Green Version]
- Hahn, S.; Jackstadt, R.; Siemens, H.; Hünten, S.; Hermeking, H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013, 32, 3079–3095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miekus, K.; Lukasiewicz, E.; Jarocha, D.; Sekula, M.; Drabik, G.; Majka, M. The decreased metastatic potential of rhabdomyosarcoma cells obtained through MET receptor downregulation and the induction of differentiation. Cell Death Dis. 2013, 4, e459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dash, P.R.; Whitley, G.S.J.; Ayling, L.J.; Johnstone, A.P.; Cartwright, J.E. Trophoblast apoptosis is inhibited by hepatocyte growth factor through the Akt and β-catenin mediated up-regulation of inducible nitric oxide synthase. Cell. Signal. 2005, 17, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Hangan, D.; Uniyal, S.; Morris, V.L.; MacDonald, I.C.; von Ballestrem, C.; Chau, T.; Schmidt, E.E.; Chambers, A.F.; Groom, A.C.; Chan, B.M. Integrin VLA-2 (alpha2beta1) function in postextravasation movement of human rhabdomyosarcoma RD cells in the liver. Cancer Res. 1996, 56, 3142–3149. [Google Scholar]
- Adamus, T.; Konieczny, P.; Sekuła, M.; Sułkowski, M.; Majka, M. The strategy of fusion genes construction determines efficient expression of introduced transcription factors. Acta Biochim. Pol. 2014, 61, 773–778. [Google Scholar] [CrossRef]
- Skupien-Rabian, B.; Jankowska, U.; Swiderska, B.; Lukasiewicz, S.; Ryszawy, D.; Dziedzicka-Wasylewska, M.; Kedracka-Krok, S. Proteomic and bioinformatic analysis of a nuclear intrinsically disordered proteome. J. Proteomics 2015, 130, 76–84. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef]
- Cote, R.G.; Griss, J.; Dianes, J.A.; Wang, R.; Wright, J.C.; van den Toorn, H.W.P.; van Breukelen, B.; Heck, A.J.R.; Hulstaert, N.; Martens, L.; et al. The PRoteomics IDEntification (PRIDE) Converter 2 Framework: An Improved Suite of Tools to Facilitate Data Submission to the PRIDE Database and the ProteomeXchange Consortium. Mol. Cell. Proteomics 2012, 11, 1682–1689. [Google Scholar] [CrossRef] [Green Version]
- Sandelin, A.; Wasserman, W.W.; Lenhard, B. ConSite: Web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004, 32, W249–W252. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davicioni, E.; Finckenstein, F.G.; Shahbazian, V.; Buckley, J.D.; Triche, T.J.; Anderson, M.J. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006, 66, 6936–6946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]












| Names | logFC | p Value | FDR |
|---|---|---|---|
| hsa-miR-1269b | −11.78 | 1.12 × 10−83 | 6.95 × 10−82 |
| hsa-miR-218-5p | −11.07 | 1.50 × 10−142 | 2.10 × 10−140 |
| hsa-miR-548h-5p | −10.81 | 5.84 × 10−61 | 1.82 × 10−59 |
| hsa-miR-452-5p | −10.73 | 3.40 × 10−174 | 1.47 × 10−171 |
| hsa-miR-224-5p | −10.68 | 1.22 × 10−40 | 1.89 × 10−39 |
| hsa-miR-3148 | 10.32 | 9.71 × 10−56 | 2.82 × 10−54 |
| hsa-miR-4652-5p | −9.78 | 7.14 × 10−43 | 1.24 × 10−41 |
| hsa-miR-1269a | −9.77 | 6.90 × 10−156 | 1.51 × 10−153 |
| hsa-miR-873-5p | −9.63 | 5.99 × 10−41 | 9.65 × 10−40 |
| hsa-miR-873-3p | −8.79 | 1.56 × 10−53 | 4.25 × 10−52 |
| hsa-miR-302a-5p | −8.07 | 8.53 × 10−45 | 1.69 × 10−43 |
| hsa-miR-105-5p | −6.72 | 2.21 × 10−61 | 7.38 × 10−60 |
| hsa-miR-139-5p | 6.53 | 2.01 × 10−65 | 8.75 × 10−64 |
| hsa-miR-199b-5p | −6.52 | 4.51 × 10−94 | 3.27 × 10−92 |
| hsa-miR-95-3p | 6.42 | 2.34 × 10−40 | 3.51 × 10−39 |
| hsa-miR-767-5p | −6.15 | 8.59 × 10−51 | 1.97 × 10−49 |
| hsa-miR-139-3p | 5.93 | 1.25 × 10−44 | 2.37 × 10−43 |
| hsa-miR-28-5p | 5.43 | 1.15 × 10−94 | 1.00 × 10−92 |
| hsa-miR-143-3p | −5.26 | 2.17 × 10−53 | 5.55 × 10−52 |
| hsa-miR-28-3p | 5.1 | 3.60 × 10−113 | 3.88 × 10−111 |
| hsa-miR-193a-5p | 4.45 | 6.18 × 10−75 | 3.36 × 10−73 |
| hsa-miR-541-3p | −4.32 | 4.72 × 10−63 | 1.71 × 10−61 |
| hsa-miR-412-5p | −4.15 | 4.11 × 10−71 | 1.99 × 10−69 |
| hsa-miR-1197 | −3.83 | 1.45 × 10−50 | 3.15 × 10−49 |
| hsa-miR-200c-3p | −3.77 | 1.50 × 10−64 | 5.95 × 10−63 |
| hsa-miR-431-5p | −3.76 | 5.00 × 10−45 | 1.04 × 10−43 |
| hsa-miR-200b-3p | −3.65 | 3.26 × 10−41 | 5.45 × 10−40 |
| hsa-miR-129-5p | −3.32 | 2.27 × 10−43 | 4.11 × 10−42 |
| hsa-miR-1180-3p | −3.27 | 4.16 × 10−51 | 1.01 × 10−49 |
| hsa-miR-486-5p | 2.74 | 9.05 × 10−40 | 1.31 × 10−38 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skrzypek, K.; Kot, M.; Konieczny, P.; Nieszporek, A.; Kusienicka, A.; Lasota, M.; Bobela, W.; Jankowska, U.; Kędracka-Krok, S.; Majka, M. SNAIL Promotes Metastatic Behavior of Rhabdomyosarcoma by Increasing EZRIN and AKT Expression and Regulating MicroRNA Networks. Cancers 2020, 12, 1870. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12071870
Skrzypek K, Kot M, Konieczny P, Nieszporek A, Kusienicka A, Lasota M, Bobela W, Jankowska U, Kędracka-Krok S, Majka M. SNAIL Promotes Metastatic Behavior of Rhabdomyosarcoma by Increasing EZRIN and AKT Expression and Regulating MicroRNA Networks. Cancers. 2020; 12(7):1870. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12071870
Chicago/Turabian StyleSkrzypek, Klaudia, Marta Kot, Paweł Konieczny, Artur Nieszporek, Anna Kusienicka, Małgorzata Lasota, Wojciech Bobela, Urszula Jankowska, Sylwia Kędracka-Krok, and Marcin Majka. 2020. "SNAIL Promotes Metastatic Behavior of Rhabdomyosarcoma by Increasing EZRIN and AKT Expression and Regulating MicroRNA Networks" Cancers 12, no. 7: 1870. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers12071870