RLIP76: A Structural and Functional Triumvirate
Abstract
:Simple Summary
Abstract
1. Introduction
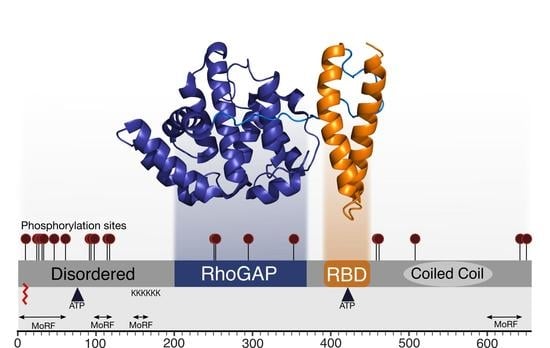
2. Domain Architecture of RLIP76
3. The RBD and the Ral-RLIP76 Complex
4. The RhoGAP Domain
5. The RLIP76 RhoGAP-RBD Didomain
6. The N-Terminal Region
7. The C-Terminal Region
8. ATP Dependent Transporter Activity
9. Localization
10. Quaternary Structure
11. Post-Translational Modifications
12. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cantor, S.B.; Urano, T.; Feig, L.A. Identification and Characterization of Ral-Binding Protein 1, a Potential Downstream Target of Ral GTPases. Mol. Cell. Biol. 1995, 15, 4578–4584. [Google Scholar] [CrossRef] [Green Version]
- Jullien-Flores, V.; Dorseuil, O.; Romero, F.; Letourneur, F.; Saragosti, S.; Berger, R.; Tavitian, A.; Gacon, G.; Camonis, J.H. Bridging Ral GTPase to Rho Pathways. RLIP76, a Ral Effector with CDC42/Rac GTPase-Activating Protein Activity. J. Biol. Chem. 1995, 270, 22473–22477. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Weinberg, R.A. A Putative Effector of Ral Has Homology to Rho/Rac GTPase Activating Proteins. Oncogene 1995, 11, 2349–2355. [Google Scholar]
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020, 80, 2969–2974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCormick, F. Sticking It to KRAS: Covalent Inhibitors Enter the Clinic. Cancer Cell 2020, 37, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Neel, N.F.; Martin, T.D.; Stratford, J.K.; Zand, T.P.; Reiner, D.J.; Der, C.J. The RalGEF-Ral Effector Signaling Network. Genes Cancer 2011, 2, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Vatsyayan, R.; Lelsani, P.C.R.; Awasthi, S.; Singhal, S.S. RLIP76: A Versatile Transporter and an Emerging Target for Cancer Therapy. Biochem. Pharmacol. 2010, 79, 1699–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajasekar, K.V.; Campbell, L.J.; Nietlispach, D.; Owen, D.; Mott, H.R. The Structure of the RLIP76 RhoGAP-Ral Binding Domain Dyad: Fixed Position of the Domains Leads to Dual Engagement of Small G Proteins at the Membrane. Structure 2013, 21, 2131–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenwick, R.B.; Campbell, L.J.; Rajasekar, K.; Prasannan, S.; Nietlispach, D.; Camonis, J.; Owen, D.; Mott, H.R. The RalB-RLIP76 (RalBP1) Complex Reveals a Novel Mode of Ral-Effector Interaction. Structure 2010, 18, 985–995. [Google Scholar] [CrossRef] [Green Version]
- Amin, E.; Jaiswal, M.; Derewenda, U.; Reis, K.; Nouri, K.; Koessmeier, K.T.; Aspenström, P.; Somlyo, A.V.; Dvorsky, R.; Ahmadian, M.R. Deciphering the Molecular and Functional Basis of RHOGAP Family Proteins: A Systematic Approach Toward Selective Inactivation of Rho Family Proteins. J. Biol. Chem. 2016, 291, 20353–20371. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Coghlan, A.; Ruan, J.; Coin, L.J.; Hériché, J.K.; Osmotherly, L.; Li, R.; Liu, T.; Zhang, Z.; Bolund, L.; et al. TreeFam: A Curated Database of Phylogenetic Trees of Animal Gene Families. Nucleic Acids Res. 2006, 34, D572–D580. [Google Scholar] [CrossRef]
- Ruan, J.; Li, H.; Chen, Z.; Coghlan, A.; Coin, L.J.; Guo, Y.; Hériché, J.K.; Hu, Y.; Kristiansen, K.; Li, R.; et al. TreeFam: 2008 Update. Nucleic Acids Res. 2007, 36, D735–D740. [Google Scholar] [CrossRef] [PubMed]
- Frische, E.W.; Pellis-van Berkel, W.; van Haaften, G.; Cuppen, E.; Plasterk, R.H.; Tijsterman, M.; Bos, J.L.; Zwartkruis, F.J.T. RAP-1 and the RAL-1/Exocyst Pathway Coordinate Hypodermal Cell Organization in Caenorhabditis Elegans. EMBO J. 2007, 26, 5083–5092. [Google Scholar] [CrossRef] [PubMed]
- Jullien-Flores, V.; Mahe, Y.; Mirey, G.; Leprince, C.; Meunier-Bisceuil, B.; Sorkin, A.; Camonis, J.H. RLIP76, an Effector of the GTPase Ral, Interacts with the AP2 Complex: Involvement of the Ral Pathway in Receptor Endocytosis. J. Cell Sci. 2000, 113, 2837–2844. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Wurtzel, J.G.T.; Goldfinger, L.E. The RLIP76 N-Terminus Binds ARNO to Regulate PI 3-Kinase, Arf6 and Rac Signaling, Cell Spreading and Migration. Biochem. Biophys. Res. Commun. 2014, 454, 560–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wurtzel, J.G.T.; Lee, S.; Singhal, S.S.; Awasthi, S.; Ginsberg, M.H.; Goldfinger, L.E. RLIP76 Regulates Arf6-Dependent Cell Spreading and Migration by Linking ARNO with Activated R-Ras at Recycling Endosomes. Biochem. Biophys. Res. Commun. 2015, 467, 785–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coon, B.G.; Burgner, J.; Camonis, J.H.; Aguilar, R.C. The Epsin Family of Endocytic Adaptors Promotes Fibrosarcoma Migration and Invasion. J. Biol. Chem. 2010, 285, 33073–33081. [Google Scholar] [CrossRef] [Green Version]
- Goldfinger, L.E.; Ptak, C.; Jeffery, E.D.; Shabanowitz, J.; Hunt, D.F.; Ginsberg, M.H. RLIP76 (RalBP1) Is an R-Ras Effector That Mediates Adhesion-Dependent Rac Activation and Cell Migration. J. Cell Biol. 2006, 174, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Urano, T.; Goi, T.; Feig, L.A. An Eps Homology (EH) Domain Protein That Binds to the Ral-GTPase Target, RalBP1. J. Biol. Chem. 1997, 272, 31230–31234. [Google Scholar] [CrossRef] [Green Version]
- Boissel, L.; Fillatre, J.; Moreau, J. Identification and Characterization of the RLIP/RALBP1 Interacting Protein Xreps1 in Xenopus Laevis Early Development. PLoS ONE 2012, 7, e33193. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Ishida, O.; Hinoi, T.; Kishida, S.; Kikuchi, A. Identification and Characterization of a Novel Protein Interacting with Ral-Binding Protein 1, a Putative Effector Protein of Ral. J. Biol. Chem. 1998, 273, 814–821. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, S.; Morinaka, K.; Koyama, S.; Ikeda, M.; Kishida, M.; Okawa, K.; Iwamatsu, A.; Kishida, S.; Kikuchi, A. Small G Protein Ral and Its Downstream Molecules Regulate Endocytosis of EGF and Insulin Receptors. EMBO J. 1999, 18, 3629–3642. [Google Scholar] [CrossRef] [PubMed]
- Rossé, C.; L’Hoste, S.; Offner, N.; Picard, A.; Camonis, J. RLIP, an Effector of the Ral GTPases, Is a Platform for Cdk1 to Phosphorylate Epsin during the Switch Off of Endocytosis in Mitosis. J. Biol. Chem. 2003, 278, 30597–30604. [Google Scholar] [CrossRef] [Green Version]
- Kariya, K.; Koyama, S.; Nakashima, S.; Oshiro, T.; Morinaka, K.; Kikuchi, A. Regulation of Complex Formation of POB1/Epsin/Adaptor Protein Complex 2 by Mitotic Phosphorylation. J. Biol. Chem. 2000, 275, 18399–18406. [Google Scholar] [CrossRef] [Green Version]
- Kashatus, D.F.; Lim, K.-H.; Brady, D.C.; Pershing, N.L.K.; Cox, A.D.; Counter, C.M. RALA and RALBP1 Regulate Mitochondrial Fission at Mitosis. Nat. Cell Biol. 2011, 13, 1108–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Mivechi, N.F. HSF-1 Interacts with Ral-Binding Protein 1 in a Stress-Responsive, Multiprotein Complex with HSP90 in Vivo. J. Biol. Chem. 2003, 278, 17299–17306. [Google Scholar] [CrossRef] [Green Version]
- Han, K.; Kim, M.-H.; Seeburg, D.; Seo, J.; Verpelli, C.; Han, S.; Chung, H.S.; Ko, J.; Lee, H.W.; Kim, K.; et al. Regulated RalBP1 Binding to RalA and PSD-95 Controls AMPA Receptor Endocytosis and LTD. PLoS Biol. 2009, 7, e1000187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, L.J.; Peppa, M.; Crabtree, M.D.; Shafiq, A.; McGough, N.F.; Mott, H.R.; Owen, D. Thermodynamic Mapping of Effector Protein Interfaces with RalA and RalB. Biochemistry 2015, 54, 1380–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurd, C.A.; Brear, P.; Revell, J.; Ross, S.; Mott, H.R.; Owen, D. Affinity Maturation of the RLIP76 Ral Binding Domain to Inform the Design of Stapled Peptides Targeting the Ral GTPases. J. Biol. Chem. 2020, 296, 100101. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.C.; Cooper, J.M.; Clayton, N.S.; Wang, C.; White, M.A.; Abell, C.; Owen, D.; Mott, H.R. Inhibition of Ral GTPases Using a Stapled Peptide Approach. J. Biol. Chem. 2016, 291, 18310–18325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, K.-H.; Brady, D.C.; Kashatus, D.F.; Ancrile, B.B.; Der, C.J.; Cox, A.D.; Counter, C.M. Aurora-A Phosphorylates, Activates, and Relocalizes the Small GTPase RalA. Mol. Cell. Biol. 2010, 30, 508–523. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neel, N.F.; Rossman, K.L.; Martin, T.D.; Hayes, T.K.; Yeh, J.J.; Der, C.J. The RalB Small GTPase Mediates Formation of Invadopodia through a GTPase-Activating Protein-Independent Function of the RalBP1/RLIP76 Effector. Mol. Cell. Biol. 2012, 32, 1374–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Vendome, J.; Shapiro, L.; Ben-Shaul, A.; Honig, B. Transforming Binding Affinities from Three Dimensions to Two with Application to Cadherin Clustering. Nature 2011, 475, 510–513. [Google Scholar] [CrossRef] [Green Version]
- Karimzadeh, F.; Primeau, M.; Mountassif, D.; Rouiller, I.; Lamarche-Vane, N. A Stretch of Polybasic Residues Mediates Cdc42 GTPase-Activating Protein (CdGAP) Binding to Phosphatidylinositol 3,4,5-Trisphosphate and Regulates Its GAP Activity. J. Biol. Chem. 2012, 287, 19610–19621. [Google Scholar] [CrossRef] [Green Version]
- Erlmann, P.; Schmid, S.; Horenkamp, F.A.; Geyer, M.; Pomorski, T.G.; Olayioye, M.A. DLC1 Activation Requires Lipid Interaction through a Polybasic Region Preceding the RhoGAP Domain. Mol. Biol. Cell 2009, 20, 4400–4411. [Google Scholar] [CrossRef]
- Mott, H.R.; Owen, D. Structure and Function of RLIP76 (RalBP1): An Intersection Point between Ras and Rho Signalling. Biochem. Soc. Trans. 2014, 42, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Mazhab-Jafari, M.T.; Marshall, C.B.; Smith, M.J.; Gasmi-Seabrook, G.M.C.; Stathopulos, P.B.; Inagaki, F.; Kay, L.E.; Neel, B.G.; Ikura, M. Oncogenic and RASopathy-Associated K-RAS Mutations Relieve Membrane-Dependent Occlusion of the Effector-Binding Site. Proc. Natl. Acad. Sci. USA 2015, 112, 6625–6630. [Google Scholar] [CrossRef] [Green Version]
- Fang, Z.; Marshall, C.B.; Nishikawa, T.; Gossert, A.D.; Jansen, J.M.; Jahnke, W.; Ikura, M. Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site. Cell Chem. Biol. 2018, 25, 1327–1336.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsubara, K.; Hinoi, T.; Koyama, S.; Kikuchi, A. The Post-Translational Modifications of Ral and Rac1 Are Important for the Action of Ral-Binding Protein 1, a Putative Effector Protein of Ral. FEBS Lett. 1997, 410, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why Are “Natively Unfolded” Proteins Unstructured under Physiologic Conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Mészáros, B.; Erdős, G.; Dosztányi, Z. IUPred2A: Context-Dependent Prediction of Protein Disorder as a Function of Redox State and Protein Binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef] [PubMed]
- Erdős, G.; Dosztányi, Z. Analyzing Protein Disorder with IUPred2A. Curr. Protoc. Bioinforma. 2020, 70, e99. [Google Scholar] [CrossRef] [Green Version]
- Romero, P.; Obradovic, Z.; Dunker, A.K. Sequence Data Analysis for Long Disordered Regions Prediction in the Calcineurin Family. Genome Inform. 1997, 8, 110–124. [Google Scholar]
- Li, X.; Romero, P.; Rani, M.; Dunker, A.K.; Obradovic, Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform. 1999, 10, 30–40. [Google Scholar]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A Meta-Predictor of Intrinsically Disordered Amino Acids. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [Green Version]
- Mohan, A.; Oldfield, C.J.; Radivojac, P.; Vacic, V.; Cortese, M.S.; Dunker, A.K.; Uversky, V.N. Analysis of Molecular Recognition Features (MoRFs). J. Mol. Biol. 2006, 362, 1043–1059. [Google Scholar] [CrossRef]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing Protein Intrinsic Disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [Green Version]
- Bah, A.; Vernon, R.M.; Siddiqui, Z.; Krzeminski, M.; Muhandiram, R.; Zhao, C.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Folding of an Intrinsically Disordered Protein by Phosphorylation as a Regulatory Switch. Nature 2015, 519, 106–109. [Google Scholar] [CrossRef]
- Giménez-Andrés, M.; Čopič, A.; Antonny, B. The Many Faces of Amphipathic Helices. Biomolecules 2018, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting Coiled Coils from Protein Sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Freddolino, P.L.; Zhang, Y. COFACTOR: Improved Protein Function Prediction by Combining Structure, Sequence and Protein–Protein Interaction Information. Nucleic Acids Res. 2017, 45, W291–W299. [Google Scholar] [CrossRef]
- Chao, L.; Lu, L.; Yang, H.; Zhu, Y.; Li, Y.; Wang, Q.; Yu, X.; Jiang, S.; Chen, Y.-H. Identification of a Human Protein-Derived HIV-1 Fusion Inhibitor Targeting the Gp41 Fusion Core Structure. PLoS ONE 2013, 8, e66156. [Google Scholar] [CrossRef] [Green Version]
- Zuo, J.; Baler, R.; Dahl, G.; Voellmy, R. Activation of the DNA-Binding Ability of Human Heat Shock Transcription Factor 1 May Involve the Transition from an Intramolecular to an Intermolecular Triple-Stranded Coiled-Coil Structure. Mol. Cell. Biol. 1994, 14, 7557–7568. [Google Scholar] [CrossRef]
- Saraste, M.; Sibbald, P.R.; Wittinghofer, A. The P-Loop--a Common Motif in ATP- and GTP-Binding Proteins. Trends Biochem. Sci. 1990, 15, 430–434. [Google Scholar] [CrossRef]
- Awasthi, S.; Cheng, J.-Z.; Singhal, S.S.; Pandya, U.; Sharma, R.; Singh, S.V.; Zimniak, P.; Awasthi, Y.C. Functional Reassembly of ATP-Dependent Xenobiotic Transport by the N- and C-Terminal Domains of RLIP76 and Identification of ATP Binding Sequences. Biochemistry 2001, 40, 4159–4168. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.E.; Saraste, M.; Runswick, M.J.; Gay, N.J. Distantly Related Sequences in the Alpha- and Beta-Subunits of ATP Synthase, Myosin, Kinases and Other ATP-Requiring Enzymes and a Common Nucleotide Binding Fold. EMBO J. 1982, 1, 945–951. [Google Scholar] [CrossRef]
- Ambudkar, S.V.; Kim, I.-W.; Xia, D.; Sauna, Z.E. The A-Loop, a Novel Conserved Aromatic Acid Subdomain Upstream of the Walker A Motif in ABC Transporters, Is Critical for ATP Binding. FEBS Lett. 2006, 580, 1049–1055. [Google Scholar] [CrossRef] [Green Version]
- Awasthi, S.; Singhal, S.S.; Singhal, J.; Nagaprashantha, L.; Li, H.; Yuan, Y.-C.; Liu, Z.; Berz, D.; Igid, H.; Green, W.C.; et al. Anticancer Activity of 2’-Hydroxyflavanone towards Lung Cancer. Oncotarget 2018, 9, 36202–36219. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Cheng, J.; Singhal, S.S.; Saini, M.K.; Pandya, U.; Pikula, S.; Bandorowicz-Pikula, J.; Singh, S.V.; Zimniak, P.; Awasthi, Y.C. Novel Function of Human RLIP76: ATP-Dependent Transport of Glutathione Conjugates and Doxorubicin. Biochemistry 2000, 39, 9327–9334. [Google Scholar] [CrossRef]
- Singhal, S.S.; Mohanty, A.; Kulkarni, P.; Horne, D.; Awasthi, S.; Salgia, R. RLIP Depletion Induces Apoptosis Associated with Inhibition of JAK2/STAT3 Signaling in Melanoma Cells. Carcinogenesis 2021. [Google Scholar] [CrossRef]
- Fillatre, J.; Delacour, D.; Van Hove, L.; Bagarre, T.; Houssin, N.; Soulika, M.; Veitia, R.A.; Moreau, J. Dynamics of the Subcellular Localization of RalBP1/RLIP through the Cell Cycle: The Role of Targeting Signals and of Protein-Protein Interactions. FASEB J. 2012, 26, 2164–2174. [Google Scholar] [CrossRef]
- Yadav, S.; Singhal, S.S.; Singhal, J.; Wickramarachchi, D.; Knutson, E.; Albrecht, T.B.; Awasthi, Y.C.; Awasthi, S. Identification of Membrane-Anchoring Domains of RLIP76 Using Deletion Mutant Analyses. Biochemistry 2004, 43, 16243–16253. [Google Scholar] [CrossRef]
- Wei, L.; Xing, P.; Su, R.; Shi, G.; Ma, Z.S.; Zou, Q. CPPred-RF: A Sequence-Based Predictor for Identifying Cell-Penetrating Peptides and Their Uptake Efficiency. J. Proteome Res. 2017, 16, 2044–2053. [Google Scholar] [CrossRef]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef] [PubMed]
- Ligeti, E.; Settleman, J. Regulation of RhoGAP Specificity by Phospholipids and Prenylation. Methods Enzymol. 2006, 406, 104–117. [Google Scholar] [PubMed]
- Mulgrew-Nesbitt, A.; Diraviyam, K.; Wang, J.; Singh, S.; Murray, P.; Li, Z.; Rogers, L.; Mirkovic, N.; Murray, D. The Role of Electrostatics in Protein–Membrane Interactions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2006, 1761, 812–826. [Google Scholar] [CrossRef]
- Das, T.; Eliezer, D. Membrane Interactions of Intrinsically Disordered Proteins: The Example of Alpha-Synuclein. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Cornish, J.; Chamberlain, S.G.; Owen, D.; Mott, H.R. Intrinsically Disordered Proteins and Membranes: A Marriage of Convenience for Cell Signalling? Biochem. Soc. Trans. 2020, 48, 2669–2689. [Google Scholar] [CrossRef]
- Kennedy, S.A.; Jarboui, M.-A.; Srihari, S.; Raso, C.; Bryan, K.; Dernayka, L.; Charitou, T.; Bernal-Llinares, M.; Herrera-Montavez, C.; Krstic, A.; et al. Extensive Rewiring of the EGFR Network in Colorectal Cancer Cells Expressing Transforming Levels of KRASG13D. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Anurag, M.; Singh, G.P.; Dash, D. Location of Disorder in Coiled Coil Proteins Is Influenced by Its Biological Role and Subcellular Localization: A GO-Based Study on Human Proteome. Mol. Biosyst. 2011, 8, 346–352. [Google Scholar] [CrossRef]
- Awasthi, S.; Singhal, S.S.; Sharma, R.; Zimniak, P.; Awasthi, Y.C. Transport of Glutathione Conjugates and Chemotherapeutic Drugs by RLIP76 (RALBP1): A Novel Link between G-Protein and Tyrosine Kinase Signaling and Drug Resistance. Int. J. Cancer 2003, 106, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Sharma, R.; Singhal, S.S.; Zimniak, P.; Awasthi, Y.C. RLIP76, a Novel Transporter Catalyzing ATP-Dependent Efflux of Xenobiotics. Drug Metab. Dispos. 2002, 30, 1300–1310. [Google Scholar] [CrossRef] [Green Version]
- Quaroni, A.; Paul, E.C. Cytocentrin Is a Ral-Binding Protein Involved in the Assembly and Function of the Mitotic Apparatus. J. Cell Sci. 1999, 112, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Maurer-Stroh, S.; Eisenhaber, B.; Eisenhaber, F. N-Terminal N-Myristoylation of Proteins: Prediction of Substrate Proteins from Amino Acid Sequence. J. Mol. Biol. 2002, 317, 541–557. [Google Scholar] [CrossRef]
- Xie, Y.; Zheng, Y.; Li, H.; Luo, X.; He, Z.; Cao, S.; Shi, Y.; Zhao, Q.; Xue, Y.; Zuo, Z.; et al. GPS-Lipid: A Robust Tool for the Prediction of Multiple Lipid Modification Sites. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Rocks, O.; Gerauer, M.; Vartak, N.; Koch, S.; Huang, Z.-P.; Pechlivanis, M.; Kuhlmann, J.; Brunsveld, L.; Chandra, A.; Ellinger, B.; et al. The Palmitoylation Machinery Is a Spatially Organizing System for Peripheral Membrane Proteins. Cell 2010, 141, 458–471. [Google Scholar] [CrossRef] [Green Version]
- Linder, M.E.; Deschenes, R.J. Palmitoylation: Policing Protein Stability and Traffic. Nat. Rev. Mol. Cell Biol. 2007, 8, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Singhal Sharad, S.; Yadav, S.; Singhal, J.; Drake, K.; Awasthi, Y.C.; Awasthi, S. The Role of PKCα and RLIP76 in Transport-mediated Doxorubicin-resistance in Lung Cancer. FEBS Lett. 2005, 579, 4635–4641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herlevsen, M.; Theodorescu, D. Mass Spectroscopic Phosphoprotein Mapping of Ral Binding Protein 1 (RalBP1/Rip1/RLIP76). Biochem. Biophys. Res. Commun. 2007, 362, 56–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tcherkezian, J.; Lamarche-Vane, N. Current Knowledge of the Large RhoGAP Family of Proteins. Biol. Cell 2007, 99, 67–86. [Google Scholar] [CrossRef]
- Ko, F.C.F.; Chan, L.-K.; Man-Fong Sze, K.; Yeung, Y.-S.; Yuk-Ting Tse, E.; Lu, P.; Yu, M.-H.; Oi-Lin Ng, I.; Yam, J.W.P. PKA-Induced Dimerization of the RhoGAP DLC1 Promotes Its Inhibition of Tumorigenesis and Metastasis. Nat. Commun. 2013, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Minoshima, Y.; Kawashima, T.; Hirose, K.; Tonozuka, Y.; Kawajiri, A.; Bao, Y.C.; Deng, X.; Tatsuka, M.; Narumiya, S.; May, W.S.; et al. Phosphorylation by Aurora B Converts MgcRacGAP to a RhoGAP during Cytokinesis. Dev. Cell 2003, 4, 549–560. [Google Scholar] [CrossRef] [Green Version]






| Site | Location in Sequence | Number of References: LTP/HTP |
|---|---|---|
| SER11 | N-terminus | 1/9 |
| THR27 | N-terminus | 1/49 |
| SER29 | N-terminus | 1/538 |
| SER30 | N-terminus | 1/103 |
| SER34 | N-terminus | 1/76 |
| SER48 | N-terminus | 1/57 |
| SER62 | N-terminus | 1/59 |
| SER92 | N-terminus | 1/41 |
| SER93 | N-terminus | 1/45 |
| SER99 | N-terminus | 1/5 |
| SER116 1 | N-terminus | 1/9 |
| SER118 1 | N-terminus | 2/3 |
| SER252 1 | RhoGAP | 1/0 |
| THR253 1 | RhoGAP | 1/0 |
| THR297 | RhoGAP | 1/0 |
| SER353 | RhoGAP | 1/0 |
| SER461 | C-terminus | 0/10 |
| SER463 | C-terminus | 1/36 |
| SER509 | C-terminus | 1/0 |
| SER645 | C-terminus | 1/14 |
| THR653 2 | C-terminus | 1/0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cornish, J.; Owen, D.; Mott, H.R. RLIP76: A Structural and Functional Triumvirate. Cancers 2021, 13, 2206. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13092206
Cornish J, Owen D, Mott HR. RLIP76: A Structural and Functional Triumvirate. Cancers. 2021; 13(9):2206. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13092206
Chicago/Turabian StyleCornish, Jasmine, Darerca Owen, and Helen R. Mott. 2021. "RLIP76: A Structural and Functional Triumvirate" Cancers 13, no. 9: 2206. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13092206