Role of the Holoenzyme PP1-SPN in the Dephosphorylation of the RB Family of Tumor Suppressors During Cell Cycle
Abstract
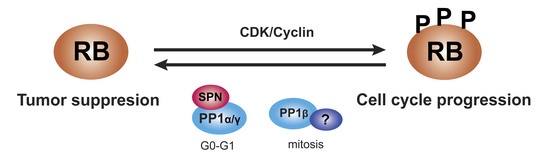
:Simple Summary
Abstract
1. Introduction
2. RB Family Proteins
Phosphorylation of Pocket Proteins
3. Protein Phosphatase 1 (PP1)
4. SPN, a PP1 Regulatory Protein
5. SPN as a Tumor Suppressor Dependent on PP1 and pRB
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berndt, N.; Dohadwala, M.; Liu, C.W. Constitutively active protein phosphatase 1alpha causes Rb-dependent G1 arrest in human cancer cells. Curr. Biol. 1997, 7, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Rubin, E.; Tamrakar, S.; Ludlow, J.W. Protein phosphatase type 1, the product of the retinoblastoma susceptibility gene, and cell cycle control. Front. Biosci. 1998, 3, D1209–D1219. [Google Scholar]
- Weinberg, R.A. The retinoblastoma protein and cell cycle control. Cell 1995, 81, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Planas-Silva, M.D.; Weinberg, R.A. The restriction point and control of cell proliferation. Curr. Opin. Cell Biol. 1997, 9, 768–772. [Google Scholar] [CrossRef]
- Bartek, J.; Bartkova, J.; Lukas, J. The retinoblastoma protein pathway and the restriction point. Curr. Opin. Cell Biol. 1996, 8, 805–814. [Google Scholar] [CrossRef]
- Schafer, K.A. The Cell Cycle: A Review. Vet. Pathol. 1998, 35, 461–478. [Google Scholar] [CrossRef]
- Krtolica, A.; Krucher, N.A.; Ludlow, J.W. Molecular analysis of selected cell cycle regulatory proteins during aerobic and hypoxic maintenance of human ovarian carcinoma cells. Br. J. Cancer 1999, 80, 1875–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berndt, N. Protein dephosphorylation and the intracellular control of the cell number. Front. Biosci. 1999, 4, D22–D42. [Google Scholar] [CrossRef] [Green Version]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef]
- Ludlow, J.W.; Nelson, D.A. Control and activity of type-1 serine/threonine protein phosphatase during the cell cycle. Semin. Cancer Biol. 1995, 6, 195–202. [Google Scholar] [CrossRef]
- Kitagawa, M.; Higashi, H.; Jung, H.K.; Suzuki-Takahashi, I.; Ikeda, M.; Tamai, K.; Kato, J.Y.; Segawa, K.; Yoshida, E.; Nishimura, S.; et al. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996, 15, 7060–7069. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.A.; Ludlow, J.W. Characterization of the mitotic phase pRb-directed protein phosphatase activity. Oncogene 1997, 14, 2407–2415. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, A.S.; Weinberg, R.A. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell Biol. 1998, 18, 753–761. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, A.S.; Weinberg, R.A. Control of the cell cycle and apoptosis. Eur. J. Cancer 1999, 35, 1886–1894. [Google Scholar] [CrossRef]
- Harbour, J.W.; Luo, R.X.; Dei Santi, A.; Postigo, A.A.; Dean, D.C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 1999, 98, 859–869. [Google Scholar] [CrossRef] [Green Version]
- Tamrakar, S.; Rubin, E.; Ludlow, J.W. Role of pRB dephosphorylation in cell cycle regulation. Front. Biosci. 2000, 5, D121–D137. [Google Scholar] [CrossRef] [Green Version]
- Classon, M.; Dyson, N. p107 and p130: Versatile proteins with interesting pockets. Exp. Cell Res. 2001, 264, 135–147. [Google Scholar] [CrossRef]
- Graña, X.; Garriga, J.; Mayol, X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene 1998, 17, 3365–3383. [Google Scholar] [CrossRef] [Green Version]
- Kolupaeva, V.; Janssens, V. PP1 and PP2A phosphatases—Cooperating partners in modulating retinoblastoma protein activation. FEBS J. 2013, 280, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, R.; D’Anneo, A.; Tesoriere, G.; Vento, R. RB1 in cancer: Different mechanisms of RB1 inactivation and alterations of pRb pathway in tumorigenesis. J. Cell Physiol. 2013, 228, 1676–1687. [Google Scholar] [CrossRef] [PubMed]
- Chinnam, M.; Goodrich, D.W. RB1, Development, and Cancer. Curr. Top. Dev. Biol. 2011, 94, 129–169. [Google Scholar]
- Guzman, F.; Fazeli, Y.; Khuu, M.; Salcido, K.; Singh, S.; Benavente, C.A. Retinoblastoma tumor suppressor protein roles in epigenetic regulation. Cancers 2020, 12, 2807. [Google Scholar] [CrossRef]
- Chen, L.; Liu, S.; Tao, Y. Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct. Target. Ther. 2020, 5, 90. [Google Scholar] [CrossRef]
- Adams, P.D. Regulation of the retinoblastoma tumor suppressor protein by cyclin/cdks. Biochim. Biophys. Acta 2001, 1471, M123–M133. [Google Scholar] [CrossRef]
- Genovese, C.; Trani, D.; Caputi, M.; Claudio, P.P. Cell cycle control and beyond: Emerging roles for the retinoblastoma gene family. Oncogene 2006, 25, 5201–5209. [Google Scholar] [CrossRef] [Green Version]
- Mittnacht, S. The retinoblastoma protein—From bench to bedside. Eur. J. Cell Biol. 2005, 84, 97–107. [Google Scholar] [CrossRef]
- Claudio, P.P.; Tonini, T.; Giordano, A. The retinoblastoma family: Twins or distant cousins? Genome Biol. 2002, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wirt, S.E.; Sage, J. p107 in the public eye: An Rb understudy and more. Cell Div. 2010, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- Garriga, J.; Limón, A.; Mayol, X.; Rane, S.G.; Albrecht, J.H.; Reddy, E.P.; Andrés, V.; Graña, X. Differential regulation of the retinoblastoma family of proteins during cell proliferation and differentiation. Biochem. J. 1998, 333, 645–654. [Google Scholar] [CrossRef] [Green Version]
- Beijersbergen, R.L.; Carlée, L.; Kerkhoven, R.M.; Bernards, R. Regulation of the retinoblastoma protein-related p107 by G1 cyclin complexes. Genes Dev. 1995, 9, 1340–1353. [Google Scholar] [CrossRef] [Green Version]
- Du, W.; Pogoriler, J. Retinoblastoma family genes. Oncogene 2006, 25, 5190–5200. [Google Scholar] [CrossRef] [Green Version]
- Mayol, X.; Grana, X. The p130 pocket protein: Keeping order at cell cycle exit/re-entrance transitions. Front. Biosci. 1998, 3, d11–d24. [Google Scholar]
- Cobrinik, D. Pocket proteins and cell cycle control. Oncogene 2005, 24, 2796–2809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Z.X.; Ginsberg, D.; Ewen, M.; Livingston, D.M. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc. Natl. Acad. Sci. USA 1996, 93, 4633–4637. [Google Scholar] [CrossRef] [Green Version]
- Dyson, N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998, 12, 2245–2262. [Google Scholar] [CrossRef] [Green Version]
- Sadasivam, S.; DeCaprio, J.A. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 2013, 13, 585–595. [Google Scholar] [CrossRef] [Green Version]
- Henley, S.A.; Dick, F.A. The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 2012, 7, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Dibling, B.; Spike, B.; Dirlam, A.; Macleod, K. New roles for the RB tumor suppressor protein. Curr. Opin. Genet. Dev. 2004, 14, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, D.L.; Morel, K.L.; Sheahan, A.V.; Richards, Z.A.; Ellis, L. The Role of RB in Prostate Cancer Progression. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 301–318. [Google Scholar]
- Dick, F.A.; Goodrich, D.W.; Sage, J.; Dyson, N.J. Non-canonical functions of the RB protein in cancer. Nat. Rev. Cancer 2018, 18, 442–451. [Google Scholar] [CrossRef]
- Classon, M.; Harlow, E. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2002, 2, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Indovina, P.; Pentimalli, F.; Conti, D.; Giordano, A. Translating RB1 predictive value in clinical cancer therapy: Are we there yet? Biochem. Pharmacol. 2019, 16, 323–334. [Google Scholar] [CrossRef]
- Mushtaq, M.; Gaza, H.V.; Kashuba, E.V. Role of the RB-Interacting Proteins in Stem Cell Biology. Adv. Cancer Res. 2016, 131, 133–157. [Google Scholar]
- Viatour, P.; Sage, J. Newly identified aspects of tumor suppression by RB. Dis. Model Mech. 2011, 4, 581–585. [Google Scholar] [CrossRef] [Green Version]
- Buchkovich, K.; Duffy, L.A.; Harlow, E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 1989, 58, 1097–1105. [Google Scholar] [CrossRef]
- Ludlow, J.W.; Shon, J.; Pipas, J.M.; Livingston, D.M.; DeCaprio, J.A. The retinoblastoma susceptibility gene product undergoes cell cycle-dependent dephosphorylation and binding to and release from SV40 large T. Cell 1990, 60, 387–396. [Google Scholar] [CrossRef]
- Hatakeyama, M.; Brill, J.A.; Fink, G.R.; Weinberg, R.A. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 1994, 8, 1759–1771. [Google Scholar] [CrossRef] [Green Version]
- Garriga, J.; Jayaraman, A.L.; Limón, A.; Jayadeva, G.; Sotillo, E.; Truongcao, M.; Patsialou, A.; Wadzinski, B.E.; Grana, X. A dynamic equilibrium between CDKs and PP2A modulates phosphorylation of pRB, p107 and p130. Cell Cycle 2004, 3, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Canhoto, A.J.; Chestukhin, A.; Litovchick, L.; DeCaprio, J.A. Phosphorylation of the retinoblastoma-related protein p130 in growth-arrested cells. Oncogene 2000, 19, 5116–5122. [Google Scholar] [CrossRef] [Green Version]
- Leng, X.; Noble, M.; Adams, P.D.; Qin, J.; Harper, J.W. Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4. Mol. Cell Biol. 2002, 22, 2242–2254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, K.; Farkas, T.; Lukas, J.; Holm, K.; Rönnstrand, L.; Bartek, J. Phosphorylation-dependent and -independent functions of p130 cooperate to evoke a sustained G1 block. EMBO J. 2001, 20, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, D.; Lukas, J.; Reed, S.I. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 2002, 16, 2946–2957. [Google Scholar] [CrossRef] [Green Version]
- Conklin, J.F.; Baker, J.; Sage, J. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat. Commun. 2012, 3, 1244. [Google Scholar] [CrossRef]
- Farkas, T.; Hansen, K.; Holm, K.; Lukas, J.; Bartek, J. Distinct phosphorylation events regulate p130- and p107-mediated repression of E2F-4. J. Biol. Chem. 2002, 277, 26741–26752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rebelo, S.; Santos, M.; Martins, F.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A.B. Protein phosphatase 1 is a key player in nuclear events. Cell Signal 2015, 27, 2589–2598. [Google Scholar] [CrossRef]
- Peti, W.; Nairn, A.C.; Page, R. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 2013, 280, 596–611. [Google Scholar] [CrossRef] [Green Version]
- Alberts, A.S.; Thorburn, A.M.; Shenolikar, S.; Mumby, M.C.; Feramisco, J.R. Regulation of cell cycle progression and nuclear affinity of the retinoblastoma protein by protein phosphatases. Proc. Natl. Acad. Sci. USA 1993, 90, 388–392. [Google Scholar] [CrossRef] [Green Version]
- Walter, G.; Mumby, M. Protein serine/threonine phosphatases and cell transformation. Biochim. Biophys. Acta 1993, 1155, 207–226. [Google Scholar] [CrossRef]
- Wera, S.; Hemmings, B.A. Serine/threonine protein phosphatases. Biochem. J. 1995, 311, 17–29. [Google Scholar] [CrossRef]
- Cohen, P.T.W. Protein phosphatase 1—Targeted in many directions. J. Cell Sci. 2002, 115, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Brautigan, D.L. Flicking the switches: Phosphorylation of serine/threonine protein phosphatases. Semin. Cancer Biol. 1995, 6, 211–217. [Google Scholar] [CrossRef]
- Dancheck, B.; Ragusa, M.J.; Allaire, M.; Nairn, A.C.; Peti, W. Molecular Investigations of the Structure and Function of the Protein Phosphatase 1:Spinophilin:Inhibitor-2 Heterotrimeric Complex. Biochemistry 2011, 50, 1238–1246. [Google Scholar] [CrossRef] [Green Version]
- Cohen, P.T.; Brewis, N.D.; Hughes, V.; Mann, D.J. Protein serine/threonine phosphatases; an expanding family. FEBS Lett. 1990, 268, 355–359. [Google Scholar] [CrossRef] [Green Version]
- Egloff, M.P.; Johnson, D.F.; Moorhead, G.; Cohen, P.T.; Cohen, P.; Barford, D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997, 16, 1876–1887. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.A.; Krucher, N.A.; Ludlow, J.W. High molecular weight protein phosphatase type 1 dephosphorylates the retinoblastoma protein. J. Biol. Chem. 1997, 272, 4528–4535. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, K.; Shima, H.; Kitagawa, Y.; Irino, S.; Sugimura, T.; Nagao, M. Identification of Members of the Protein Phosphatase 1 Gene Family in the Rat and Enhanced Expression of Protein Phosphatase 1α Gene in Rat Hepatocellular Carcinomas. Jpn. J. Cancer Res. 1990, 81, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Terry-Lorenzo, R.T.; Carmody, L.C.; Voltz, J.W.; Connor, J.H.; Li, S.; Donelson Smith, F.; Milgram, S.L.; Colbran, R.J.; Shenolikar, S. The neuronal actin-binding proteins, neurabin I and neurabin II, recruit specific isoforms of protein phosphatase-1 catalytic subunits. J. Biol. Chem. 2002, 277, 27716–27724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirschi, A.; Cecchini, M.; Steinhardt, R.C.; Schamber, M.R.; Dick, F.A.; Rubin, S.M. An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat. Struct. Mol. Biol. 2010, 17, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Felgueiras, J.; Jerónimo, C.; Fardilha, M. Protein phosphatase 1 in tumorigenesis: Is it worth a closer look? Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188433. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.J.; Dancheck, B.; Critton, D.A.; Nairn, A.C.; Page, R.; Peti, W. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat. Struct. Mol. Biol. 2010, 17, 459–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragusa, M.J.; Allaire, M.; Nairn, A.C.; Page, R.; Peti, W. Flexibility in the PP1:spinophilin holoenzyme. FEBS Lett. 2011, 585, 36–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heroes, E.; Lesage, B.; Görnemann, J.; Beullens, M.; Van Meervelt, L.; Bollen, M. The PP1 binding code: A molecular-lego strategy that governs specificity. FEBS J. 2013, 280, 584–595. [Google Scholar] [CrossRef]
- Köhn, M. Turn and Face the Strange: § A New View on Phosphatases. ACS Cent. Sci. 2020, 6, 467–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludlow, J.W.; Glendening, C.L.; Livingston, D.M.; DeCarprio, J.A. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol. Cell Biol. 1993, 13, 367–372. [Google Scholar] [CrossRef] [Green Version]
- Tamrakar, S.; Mittnacht, S.; Ludlow, J.W. Binding of select forms of pRB to protein phosphatase type 1 independent of catalytic activity. Oncogene 1999, 18, 7803–7809. [Google Scholar] [CrossRef] [Green Version]
- Durfee, T.; Becherer, K.; Chen, P.L.; Yeh, S.H.; Yang, Y.; Kilburn, A.E.; Lee, W.H.; Elledge, S.J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993, 7, 555–569. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.W.; Wang, R.H.; Dohadwala, M.; Schönthal, A.H.; Villa-Moruzzi, E.; Berndt, N. Inhibitory phosphorylation of PP1alpha catalytic subunit during the G(1)/S transition. J. Biol. Chem. 1999, 274, 29470–29475. [Google Scholar] [CrossRef] [Green Version]
- Verdugo-Sivianes, E.M.; Rojas, A.M.; Muñoz-Galván, S.; Otero-Albiol, D.; Carnero, A. Mutation of SPINOPHILIN (PPP1R9B) found in human tumors promotes the tumorigenic and stemness properties of cells. Theranostics 2021, 11, 3452–3471. [Google Scholar] [CrossRef]
- Puntoni, F.; Villa-Moruzzi, E. Protein phosphatase-1 alpha, gamma 1, and delta: Changes in phosphorylation and activity in mitotic HeLa cells and in cells released from the mitotic block. Arch. Biochem. Biophys. 1997, 340, 177–184. [Google Scholar] [CrossRef]
- Puntoni, F.; Villa-Moruzzi, E. Association of protein phosphatase-1delta with the retinoblastoma protein and reversible phosphatase activation in mitotic HeLa cells and in cells released from mitosis. Biochem. Biophys. Res. Commun. 1997, 235, 704–708. [Google Scholar] [CrossRef]
- Dohadwala, M.; Silva, E.F.D.C.E.; Hall, F.L.; Williams, R.T.; Carbonaro-Hall, D.A.; Nairn, A.C.; Greengard, P.; Berndt, N. Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc. Natl. Acad. Sci. USA 1994, 91, 6408–6412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.W.Y.; Wang, R.-H.; Berndt, N. Protein phosphatase 1α activity prevents oncogenic transformation. Mol. Carcinog. 2006, 45, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Q.; Guo, J.Y.; Tang, W.; Yang, C.-S.; Freel, C.D.; Chen, C.; Nairn, A.C.; Kornbluth, S. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat. Cell Biol. 2009, 11, 644–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, E.; Mittnacht, S.; Villa-Moruzzi, E.; Ludlow, J.W. Site-specific and temporally-regulated retinoblastoma protein dephosphorylation by protein phosphatase type 1. Oncogene 2001, 20, 3776–3785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, P.B.; Ouimet, C.C.; Greengard, P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc. Natl. Acad. Sci. USA 1997, 94, 9956–9961. [Google Scholar] [CrossRef] [Green Version]
- Satoh, A.; Nakanishi, H.; Obaishi, H.; Wada, M.; Takahashi, K.; Satoh, K.; Hirao, K.; Nishioka, H.; Hata, Y.; Mizoguchi, A.; et al. Neurabin-II/Spinophilin. An Actin Filament-Binding Protein with One Pdz Domain Localized at Cadherin-Based Cell-Cell Adhesion Sites. J. Biol. Chem. 1998, 273, 3470–3475. [Google Scholar] [CrossRef] [Green Version]
- Ferrer, I.; Blanco-Aparicio, C.; Peregrino, S.; Cañamero, M.; Fominaya, J.; Cecilia, Y.; Lleonart, M.; Hernandez-Losa, J.; Ramon y Cajal, S.; Carnero, A. Spinophilin acts as a tumor suppressor by regulating Rb phosphorylation. Cell Cycle 2011, 10, 2751–2762. [Google Scholar] [CrossRef] [Green Version]
- Carnero, A. Spinophilin: A new tumor suppressor at 17q21. Curr. Mol. Med. 2012, 12, 528–535. [Google Scholar] [CrossRef] [Green Version]
- Sarrouilhe, D.; di Tommaso, A.; Métayé, T.; Ladeveze, V. Spinophilin: From partners to functions. Biochimie 2006, 88, 1099–1113. [Google Scholar] [CrossRef]
- Ferrer, I.; Peregrino, S.; Cañamero, M.; Cecilia, Y.; Blanco-Aparicio, C.; Carnero, A. Spinophilin loss contributes to tumorigenesis in vivo. Cell Cycle 2011, 10, 1948–1955. [Google Scholar] [CrossRef] [Green Version]
- Ferrer, I.; Verdugo-Sivianes, E.M.; Castilla, M.A.; Melendez, R.; Marin, J.J.; Muňoz-Galvan, S.; Lopez-Guerra, J.L.; Vieites, B.; Ortiz-Gordillo, M.J.; De Leon, J.M.; et al. Loss of the tumor suppressor spinophilin (PPP1R9B) increases the cancer stem cell population in breast tumors. Oncogene 2016, 35, 2777–2788. [Google Scholar] [CrossRef]
- Estevez-Garcia, P.; Lopez-Calderero, I.; Molina-Pinelo, S.; Muñoz-Galvan, S.; Salinas, A.; Gomez-Izquierdo, L.; Lucena-Cacace, A.; Felipe-Abrio, B.; Paz-Ares, L.; Garcia-Carbonero, R.; et al. Spinophilin loss correlates with poor patient prognosis in advanced stages of colon carcinoma. Clin. Cancer Res. 2013, 19, 3925–3935. [Google Scholar] [CrossRef] [Green Version]
- Molina-Pinelo, S.; Ferrer, I.; Blanco-Aparicio, C.; Peregrino, S.; Pastor, M.D.; Alvarez-Vega, J.; Suarez, R.; Verge, M.; Marin, J.J.; Hernández-Losa, J.; et al. Down-regulation of spinophilin in lung tumours contributes to tumourigenesis. J. Pathol. 2011, 225, 73–82. [Google Scholar] [CrossRef]
- Verdugo-Sivianes, E.M.; Navas, L.; Molina-Pinelo, S.; Ferrer, I.; Quintanal-Villalonga, A.; Peinado, J.; Garcia-Heredia, J.M.; Felipe-Abrio, B.; Muñoz-Galvan, S.; Marin, J.J.; et al. Coordinated downregulation of Spinophilin and the catalytic subunits of PP1, PPP1CA/B/C, contributes to a worse prognosis in lung cancer. Oncotarget 2017, 8, 105196–105210. [Google Scholar] [CrossRef]
- Schwarzenbacher, D.; Stiegelbauer, V.; Deutsch, A.; Ress, A.L.; Aigelsreiter, A.; Schauer, S.; Wagner, K.; Langsenlehner, T.; Resel, M.; Gerger, A.; et al. Low spinophilin expression enhances aggressive biological behavior of breast cancer. Oncotarget 2015, 6, 11191–11202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, A.P.; Smith, F.D.; VanDongen, H.M.; VanDongen, A.M.J.; Milgram, S.L. The identification of a second actin-binding region in spinophilin/neurabin II. Mol. Brain Res. 2004, 124, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Carmody, L.C.; Baucum, A.J.; Bass, M.A.; Colbran, R.J. Selective targeting of the γ1 isoform of protein phosphatase 1 to F-actin in intact cells requires multiple domains in spinophilin and neurabin. FASEB J. 2008, 22, 1660–1671. [Google Scholar] [CrossRef] [Green Version]
- Vivo, M.; Calogero, R.A.; Sansone, F.; Calabrò, V.; Parisi, T.; Borrelli, L.; Saviozzi, S.; La Mantia, G. The Human Tumor Suppressor ARF Interacts with Spinophilin/Neurabin II, a Type 1 Protein-phosphatase-binding Protein. J. Biol. Chem. 2001, 276, 14161–14169. [Google Scholar] [CrossRef] [Green Version]
- Fisher, L.A.; Wang, L.; Wu, L.; Peng, A. Phosphatase 1 nuclear targeting subunit is an essential regulator of M-phase entry, maintenance, and exit. J. Biol. Chem. 2014, 289, 23745–23752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossman, S.D.; Futter, M.; Snyder, G.L.; Allen, P.B.; Nairn, A.C.; Greengard, P.; Hsieh-Wilson, L.C. Spinophilin is phosphorylated by Ca2+/calmodulin-dependent protein kinase II resulting in regulation of its binding to F-actin. J. Neurochem. 2004, 90, 317–324. [Google Scholar] [CrossRef]
- Futter, M.; Uematsu, K.; Bullock, S.A.; Kim, Y.; Hemmings, H.C.; Nishi, A.; Greengard, P.; Nairn, A.C. Phosphorylation of spinophilin by ERK and cyclin-dependent PK 5 (Cdk5). Proc. Natl. Acad. Sci. USA 2005, 102, 3489–3494. [Google Scholar] [CrossRef] [Green Version]
- Hsieh-Wilson, L.C.; Benfenati, F.; Snyder, G.L.; Allen, P.B.; Nairn, A.C.; Greengard, P. Phosphorylation of spinophilin modulates its interaction with actin filaments. J. Biol. Chem. 2003, 278, 1186–1194. [Google Scholar] [CrossRef] [Green Version]
- Edler, M.C.; Salek, A.B.; Watkins, D.S.; Kaur, H.; Morris, C.W.; Yamamoto, B.K.; Baucum, A.J. Mechanisms Regulating the Association of Protein Phosphatase 1 with Spinophilin and Neurabin. ACS Chem. Neurosci. 2018, 9, 2701–2712. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, M.; Prokscha, A.; Eichele, G. Neurabin II mediates doublecortin-dephosphorylation on actin filaments. Biochem. Biophys. Res. Commun. 2006, 343, 839–847. [Google Scholar] [CrossRef]
- Shmueli, A.; Gdalyahu, A.; Sapoznik, S.; Sapir, T.; Tsukada, M.; Reiner, O. Site-specific dephosphorylation of doublecortin (DCX) by protein phosphatase 1 (PP1). Mol. Cell Neurosci. 2006, 32, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Bielas, S.L.; Serneo, F.F.; Chechlacz, M.; Deerinck, T.J.; Perkins, G.A.; Allen, P.B.; Ellisman, M.H.; Gleeson, J.G. Spinophilin Facilitates PP1-Mediated Dephosphorylation of PSer297 Doublecortin in Microtubule Bundling at the Axonal Wrist. Cell 2007, 129, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Abujiang, P.; Mori, T.J.; Takahashi, T.; Tanaka, F.; Kasyu, I.; Hitomi, S.; Hiai, H. Loss of heterozygosity (LOH) at 17q and 14q in human lung cancers. Oncogene 1998, 17, 3029–3033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.A.; Easton, D.F.; Ford, D.; Peto, J.; Anderson, K.; Averill, D.; Stratton, M.; Ponder, M.; Pye, C.; Ponder, B.A. Genetic heterogeneity and localization of a familial breast-ovarian cancer gene on chromosome 17q12-q21. Am. J Hum. Genet. 1993, 52, 767–776. [Google Scholar]
- Caduff, R.F.; Svoboda-Newman, S.M.; Ferguson, A.W.; Frank, T.S. Comparison of alterations of chromosome 17 in carcinoma of the ovary and of the breast. Virchows Arch. 1999, 434, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Porter, D.E.; Steel, C.M.; Cohen, B.B.; Wallace, M.R.; Carothers, A.; Chetty, U.; Carter, D.C. Genetic linkage analysis applied to unaffected women from families with breast cancer can discriminate high- from low-risk individuals. Br. J. Surg. 1993, 80, 1381–1385. [Google Scholar] [CrossRef]
- Porter, D.E.; Cohen, B.B.; Wallace, M.R.; Smyth, E.; Chetty, U.; Dixon, J.M.; Steel, C.M.; Carter, D.C. Breast cancer incidence, penetrance and survival in probable carriers of BRCA1 gene mutation in families linked to BRCA1 on chromosome 17q12-21. Br. J. Surg. 1994, 81, 1512–1515. [Google Scholar] [CrossRef] [PubMed]
- Cohen, B.B.; Porter, D.E.; Wallace, M.R.; Carothers, A.; Steel, C.M. Linkage of a major breast cancer gene to chromosome 17q12-21: Results from 15 Edinburgh families. Am. J. Hum. Genet. 1993, 52, 723–729. [Google Scholar]
- Easton, D.F.; Bishop, D.T.; Ford, D.; Crockford, G.P. Genetic linkage analysis in familial breast and ovarian cancer: Results from 214 families. The Breast Cancer Linkage Consortium. Am. J. Hum Genet. 1993, 52, 678–701. [Google Scholar]
- Aigelsreiter, A.; Ress, A.L.; Bettermann, K.; Schauer, S.; Koller, K.; Eisner, F.; Kiesslich, T.; Stojakovic, T.; Samonigg, H.; Kornprat, P.; et al. Low expression of the putative tumour suppressor spinophilin is associated with higher proliferative activity and poor prognosis in patients with hepatocellular carcinoma. Br. J. Cancer 2013, 108, 1830–1837. [Google Scholar] [CrossRef] [PubMed]
- Aigelsreiter, A.M.; Aigelsreiter, A.; Wehrschuetz, M.; Ress, A.L.; Koller, K.; Salzwimmer, M.; Gerger, A.; Schauer, S.; Bauernhofer, T.; Pichler, M. Loss of the putative tumor suppressor protein spinophilin is associated with poor prognosis in head and neck cancer. Hum. Pathol. 2014, 45, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Allen, P.B.; Zachariou, V.; Svenningsson, P.; Lepore, A.C.; Centonze, D.; Costa, C.; Rossi, S.; Bender, G.; Chen, G.; Feng, J.; et al. Distinct roles for spinophilin and neurabin in dopamine-mediated plasticity. Neuroscience 2006, 140, 897–911. [Google Scholar] [CrossRef]
- Salek, A.B.; Edler, M.C.; McBride, J.P.; Baucum, A.J. Spinophilin regulates phosphorylation and interactions of the GluN2B subunit of the N-methyl-d-aspartate receptor. J. Neurochem. 2019, 151, 185–203. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Yan, Z.; Ferreira, A.; Tomizawa, K.; Liauw, J.A.; Zhuo, M.; Allen, P.B.; Ouimet, C.C.; Greengard, P. Spinophilin regulates the formation and function of dendritic spines. Proc. Natl. Acad. Sci. USA 2000, 97, 9287–9292. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Deng, T.; Jones, R.; Li, H.; Herschkowitz, J.I.; Liu, J.C.; Weigman, V.J.; Tsao, M.-S.; Lane, T.F.; Perou, C.M.; et al. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J. Clin. Investig. 2010, 120, 3296–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnero, A.; Garcia-Mayea, Y.; Mir, C.; Lorente, J.; Rubio, I.T.; LLeonart, M.E. The cancer stem-cell signaling network and resistance to therapy. Cancer Treat Rev. 2016, 49, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Kareta, M.S.; Gorges, L.L.; Hafeez, S.; Benayoun, B.A.; Marro, S.; Zmoos, A.F.; Cecchini, M.J.; Spacek, D.; Batista, L.F.; O’Brien, M.; et al. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell 2015, 16, 39–50. [Google Scholar] [CrossRef] [Green Version]
- She, S.; Wei, Q.; Kang, B.; Wang, Y.-J. Cell cycle and pluripotency: Convergence on octamer-binding transcription factor 4. Mol. Med. Rep. 2017, 16, 6459–6466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.-L.; Chiou, S.-H.; Wu, C.-W. Targeting cancer stem cells: Emerging role of Nanog transcription factor. OncoTargets Ther. 2013, 6, 1207–1220. [Google Scholar]
- Zhang, W.; Sui, Y.; Ni, J.; Yang, T. Insights into the Nanog gene: A propeller for stemness in primitive stem cells. Int. J. Biol. Sci. 2016, 12, 1372–1381. [Google Scholar] [CrossRef] [Green Version]
- Schoeftner, S.; Scarola, M.; Comisso, E.; Schneider, C.; Benetti, R. An Oct4-pRb Axis, Controlled by MiR-335, Integrates Stem Cell Self-Renewal and Cell Cycle Control. Stem Cells 2013, 31, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Comisso, E.; Scarola, M.; Rosso, M.; Piazza, S.; Marzinotto, S.; Ciani, Y.; Orsaria, M.; Mariuzzi, L.; Schneider, C.; Schoeftner, S.; et al. OCT4 controls mitotic stability and inactivates the RB tumor suppressor pathway to enhance ovarian cancer aggressiveness. Oncogene 2017, 36, 4253–4266. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verdugo-Sivianes, E.M.; Carnero, A. Role of the Holoenzyme PP1-SPN in the Dephosphorylation of the RB Family of Tumor Suppressors During Cell Cycle. Cancers 2021, 13, 2226. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13092226
Verdugo-Sivianes EM, Carnero A. Role of the Holoenzyme PP1-SPN in the Dephosphorylation of the RB Family of Tumor Suppressors During Cell Cycle. Cancers. 2021; 13(9):2226. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13092226
Chicago/Turabian StyleVerdugo-Sivianes, Eva M., and Amancio Carnero. 2021. "Role of the Holoenzyme PP1-SPN in the Dephosphorylation of the RB Family of Tumor Suppressors During Cell Cycle" Cancers 13, no. 9: 2226. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers13092226