Isomorphous Crystals Formed by the Similar Supramolecular Motifs in Sorafenib Hydrochloride and Regorafenib Hydrochloride Salts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis and Crystallization
2.2. Single-Crystal X-ray Diffraction (SC-XRD)
2.3. Hirshfeld Surface and Fingerprint Plot Calculation
2.4. Thermal Analysis
2.5. Solubility and Dissolution Measurement
3. Results
3.1. Isomorphous Phenomenon
3.2. Crystal Structure
3.3. Hirshfeld Surface and Non-Covalent Interactions in Crystal Structure Analysis
3.4. Thermal Analyses
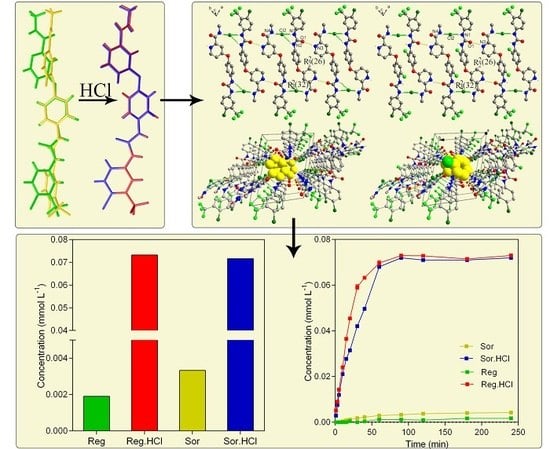
3.5. Solubility and Dissolution Rate of (I) and (II)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Poovi, G.; Damodharan, N. Lipid nanoparticles: A challenging approach for oral delivery of BCS Class-II drugs. Future J. Pharm. Sci. 2018, 4, 191–205. [Google Scholar] [CrossRef]
- Suresh, K.; Mannava, M.K.; Nangia, A. Cocrystals and alloys of nitazoxanide: Enhanced pharmacokinetics. Chem. Commun. 2016, 52, 4223–4226. [Google Scholar] [CrossRef] [PubMed]
- Rajput, L. Stable Crystalline Salts of Haloperidol: A Highly Water-Soluble Mesylate Salt. Cryst. Growth Des. 2014, 14, 5196–5205. [Google Scholar] [CrossRef]
- Zhou, X.; Hu, X.; Wu, S.; Ye, J.; Sun, M.; Gu, J.; Zhu, J.; Zhang, Z. Structures and physicochemical properties of vortioxetine salts. Acta Crystallogr. B Struct. Sci. Cryst. Eng. Mater. 2016, 72, 723–732. [Google Scholar] [CrossRef]
- Stahl, P.H.; Wermuth, C.G. Handbook of Pharmaceutical Salts: Properties, selection and use. Chem. Int. 2002, 24, 21. [Google Scholar] [CrossRef]
- Alfazari, A.S.; Almarzooqi, S.; Albawardi, A.; Saraswathiamma, D.; Abdul-Kader, H.M.; Shaban, S.; Souid, A.-K. Modulation of Cardiomyocyte and Hepatocyte Bioenergetics by Biguanides. J. Clin. Toxicol. 2014, 4, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Takimoto, C.H.; Awada, A. Safety and anti-tumor activity of sorafenib (Nexavar) in combination with other anti-cancer agents: A review of clinical trials. Cancer Chemother Pharm. 2008, 61, 535–548. [Google Scholar] [CrossRef]
- Iyer, R.; Fetterly, G.; Lugade, A.; Thanavala, Y.; Sorafenib. A clinical and pharmacologic review. Expert Opin. Pharm. 2010, 11, 1943–1955. [Google Scholar] [CrossRef]
- Sun, M.Y.; Wu, S.X.; Zhou, X.B.; Gu, J.M.; Hu, X.R. Comparison of the crystal structures of the potent anticancer and anti-angiogenic agent regorafenib and its monohydrate. Acta Crystallogr. Sect. C Struct. Chem. 2016, 72, 291–296. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Zhang, J.; Zhang, Y.; Li, Y.; Liu, Z.; Li, Q.; Luo, M.; Liang, R.; Ye, J. Molecular targeted and immune checkpoint therapy for advanced hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 447. [Google Scholar] [CrossRef]
- Giglio, V.; Viale, M.; Bertone, V.; Maric, I.; Vaccarone, R.; Vecchio, G. Cyclodextrin polymers as nanocarriers for sorafenib. Invest. New Drugs 2018, 36, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, E.; Schellens, J.H.; Beijnen, J.H.; Nuijen, B. Inventory of oral anticancer agents: Pharmaceutical formulation aspects with focus on the solid dispersion technique. Cancer Treat. Rev. 2016, 50, 247–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gidwani, R.M.; Wakchaure, V.S.; Striegel, H.G.; Albrecht, W. Polymorphorphs of 4-[4-[[4-chloro-3-(trifluoromethyl)phenyl]carbamoylamino]phenoxy]-n-methyl-pyridine-2-carboxamide, WO Patent 2010/142678 A2, 16 December 2010. [Google Scholar]
- Jaryal, J.S.; Sathyanarayana, S.; Thaper, R.K.; Prasad, M. Polymorphs of Sorafenib Acid Addition Salts. US Patent 2012/0264789 A1, 18 October 2012. [Google Scholar]
- Jacques, D.; Stephen, B.; Bernd, R.; Scott, W. Fluoro Substituted Omege-Carboxyaryl Diphenyl Urea for the Treatment and Prevention of Diseases and Conditions. WO Patent 2005/009961 A2, 3 February 2005. [Google Scholar]
- Yang, P.; Qin, C.; Du, S.; Jia, L.; Qin, Y.; Gong, J.; Wu, S. Crystal Structure, Stability and Desolvation of the Solvates of Sorafenib Tosylate. Crystals 2019, 9, 367. [Google Scholar] [CrossRef] [Green Version]
- Bruker AXS announces novel APEX(TM) DUO. The most versatile system for small molecule X-ray crystallography. Anti-Corros. Methods Mater. 2007, 54, 375.
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H.; OLEX2. A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT - integrated space-group and crystal-structure determination. Acta Crystallogr. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Blatov, V.A. Multipurpose crystallochemical analysis with the program package TOPOS. IUCr Compcomm Newsl. 2006, 7, 4–38. [Google Scholar]
- Baburin, I.A.; Blatov, V.A. Three-dimensional hydrogen-bonded frameworks in organic crystals: A topological study. Acta Crystallogr. Sect. B Struct. Sci. 2007, 63, 791–802. [Google Scholar] [CrossRef]
- Kálmán, A.; Párkányi, L.; Argay, G. Classification of the isostructurality of organic molecules in the crystalline state. Acta Crystallogr. Sect. B 1993, 49, 1039–1049. [Google Scholar] [CrossRef]
- Fábián, L.; Kálmán, A. Volumetric measure of isostructurality. Acta Crystallogr. Sect. B 1999, 55, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- CrystalExplorer; Version 3.1; University of Western Australia: Crawley, AU, Australia, 2005–2013.
- Nath, N.K.; Nangia, A. Isomorphous Crystals by Chloro–Methyl Exchange in Polymorphic Fuchsones. Cryst. Growth Des. 2012, 12, 5411–5425. [Google Scholar] [CrossRef]
- Simon, K.; Harmat, V.; Török, Z.; Böcskei, Z.; Hermecz, I. Isostructural Metabolites of Two Anti-Parkinson Drugs. Acta Crystallogr. Sect. C 1998, 54, 811–813. [Google Scholar] [CrossRef]
- Thakur, A.; Kumar, D.; Thipparaboina, R.; Shastri, N.R. Exploration of crystal simulation potential by fluconazole isomorphism and its application in improvement of pharmaceutical properties. J. Cryst. Growth 2014, 406, 18–25. [Google Scholar] [CrossRef]
- Bacchi, A.; Carcelli, M.; Pelizzi, G. Sampling rifamycin conformational variety by cruising through crystal forms: Implications for polymorph screening and for biological models. New J. Chem. 2008, 32, 1725–1735. [Google Scholar] [CrossRef]
- Dey, D.; Chopra, D. Evaluation of the Role of Isostructurality in Fluorinated Phenyl Benzoates. Cryst. Growth Des. 2017, 17, 5117–5128. [Google Scholar] [CrossRef]
- Ebenezer, S.; Muthiah, P.T.; Butcher, R.J. Design of a Series of Isostructural Co-Crystals with Aminopyrimidines: Isostructurality through Chloro/Methyl Exchange and Studies on Supramolecular Architectures. Cryst. Growth Des. 2011, 11, 3579–3592. [Google Scholar] [CrossRef]
- Capacci-Daniel, C.; Dehghan, S.; Wurster, V.M.; Basile, J.A.; Hiremath, R.; Sarjeant, A.A.; Swift, J.A. Halogen/methyl exchange in a series of isostructural 1,3-bis(m-dihalophenyl)ureas. CrystEngComm 2008, 10, 1875–1880. [Google Scholar] [CrossRef]
- Ravikumar, K.; Sridhar, B.; Bhujanga Rao, A.K.S.; Pulla Reddy, M. Sorafenib and its tosylate salt: A multikinase inhibitor for treating cancer. Acta Crystallogr. Sect. C 2011, 67, o29–o32. [Google Scholar] [CrossRef]
- Spackman, M.A.; Byrom, P.G. A novel definition of a molecule in a crystal. Chem. Phys. Lett. 1997, 267, 215–220. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J.; Jayatilaka, D. Electrostatic potentials mapped on Hirshfeld surfaces provide direct insight into intermolecular interactions in crystals. CrystEngComm 2008, 10, 377–388. [Google Scholar] [CrossRef]
- Angira, D.; Shaik, A.; Kirubakaran, S.; Thiruvenkatam, V. Exploring a solvated dimer of Gefitinib: A quantitative analysis. Acta Crystallogr. C Struct. Chem. 2018, 74, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, Z.; Chen, Y.; Lu, J.; Li, Y.; Wang, S.; Wu, G.; Qian, F. Improving Oral Bioavailability of Sorafenib by Optimizing the “Spring” and “Parachute” Based on Molecular Interaction Mechanisms. Mol. Pharm. 2016, 13, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Etter, M.C. Encoding and decoding hydrogen-bond patterns of organic compounds. Acc. Chem. Res. 1990, 23, 120–126. [Google Scholar] [CrossRef]
- Etter, M.C.; MacDonald, J.C.; Bernstein, J. Graph-set analysis of hydrogen-bond patterns in organic crystals. Acta Crystallogr. Sect. B 1990, 46, 256–262. [Google Scholar] [CrossRef]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.L. Patterns in Hydrogen Bonding: Functionality and Graph Set Analysis in Crystals. Angew. Chem. Int. Ed. Engl. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Thiagarajan, D.; Mehta, D.P. Faster Algorithms for Isomer Network Generation. J. Chem. Inf. Modeling 2016, 56, 2310–2319. [Google Scholar] [CrossRef]










| Parameter | (I) | (II) |
|---|---|---|
| Chemical formula | C21H17ClF3N4O3·Cl | C21H16ClF4N4O3·Cl |
| Crystal system, space group | Triclinic, P-1 | Triclinic, P-1 |
| Temperature (K) | 170 | 170 |
| a, b, c (nm) | 0.94153 (2), 1.05000 (3), 1.24470 (3) | 0.9458 (2), 1.0488 (3), 1.2538 (3) |
| α, β, γ (°) | 75.089 (1), 86.160 (1), 66.078 (1) | 75.282 (11), 86.190 (13), 66.055 (10) |
| V (nm3) | 1.08597 (5) | 1.0984 (5) |
| Z | 2 | 2 |
| µ (Mo Kα, mm−1) | 0.36 | 0.36 |
| Rint | 0.050 | 0.038 |
| R[F2 > 2σ(F2)], wR(F2), S | 0.046, 0.120, 1.03 | 0.053, 0.156, 1.02 |
| No. of reflections | 6337 | 16140 |
| The Cambridge Crystallographic Data Centre (CCDC) number | 1948352 | 1948353 |
| D–H···A | D–H | H···A | D···A | D–H···A |
|---|---|---|---|---|
| Sor.HCl (I) | ||||
| N3–H3···O1i | 0.088 | 0.198 | 0.283 | 165.0 |
| N1–H1···Cl2 | 0.088 | 0.230 | 0.315 | 162.9 |
| N2–H2···Cl2 | 0.088 | 0.247 | 0.330 | 157.0 |
| N4–H4···Cl2ii | 0.088 | 0.231 | 0.315 | 160.0 |
| C6–H6···O3 | 0.095 | 0.233 | 0.293 | 120.4 |
| Reg.HCl (II) | ||||
| N3–H3···O1i | 0.089 | 0.200 | 0.285 | 158.9 |
| N1–H1···Cl2 | 0.088 | 0.226 | 0.313 | 170.0 |
| N2–H2···Cl2 | 0.088 | 0.252 | 0.333 | 155.2 |
| N4–H4···Cl2ii | 0.088 | 0.232 | 0.316 | 158.6 |
| C6–H6…O3 | 0.095 | 0.234 | 0.294 | 120.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phan, C.U.; Shen, J.; Liu, J.; Mao, J.; Hu, X.; Tang, G. Isomorphous Crystals Formed by the Similar Supramolecular Motifs in Sorafenib Hydrochloride and Regorafenib Hydrochloride Salts. Crystals 2019, 9, 649. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst9120649
Phan CU, Shen J, Liu J, Mao J, Hu X, Tang G. Isomorphous Crystals Formed by the Similar Supramolecular Motifs in Sorafenib Hydrochloride and Regorafenib Hydrochloride Salts. Crystals. 2019; 9(12):649. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst9120649
Chicago/Turabian StylePhan, Chi Uyen, Jie Shen, Jiyong Liu, Jianming Mao, Xiurong Hu, and Guping Tang. 2019. "Isomorphous Crystals Formed by the Similar Supramolecular Motifs in Sorafenib Hydrochloride and Regorafenib Hydrochloride Salts" Crystals 9, no. 12: 649. https://0-doi-org.brum.beds.ac.uk/10.3390/cryst9120649