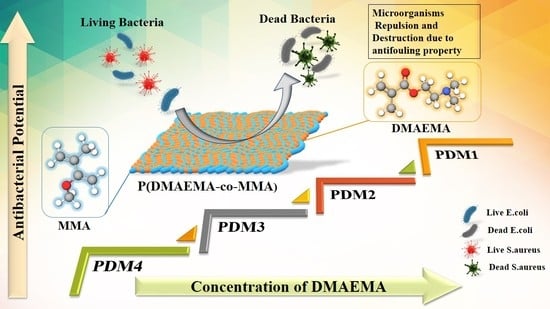
Antibacterial Amphiphilic Copolymers of Dimethylamino Ethyl Methacrylate and Methyl Methacrylate to Control Biofilm Adhesion for Antifouling Applications
Abstract
:1. Introduction
2. Materials and Methodology
2.1. Materials
2.2. Synthesis of Copolymer P(DMAEMA-co-MMA)
2.3. Characterization
2.4. Antibacterial Bioassay
2.5. Biofilm Formation Test
3. Results and Discussion
3.1. FTIR Analysis
3.2. 1HNMR Analysis
3.3. GPC Results
3.4. Antibacterial Bioassay
3.5. Biofilm Adhesion Studies by SEM
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Xue, Y.; Xiao, H.; Zhang, Y. Antimicrobial polymeric materials with quaternary ammonium and phosphonium salts. Int. J. Mol. Sci. 2015, 16, 3626–3655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tejero, R.; Gutiérrez, B.; López, D.; López-Fabal, F.; Gómez-Garcés, J.L.; Muñoz-Bonilla, A.; Fernández-García, M. Tailoring macromolecular structure of cationic polymers towards efficient contact active antimicrobial surfaces. Polymers (Basel) 2018, 10, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.; Yang, C.; Huang, S.; Chen, C.; Lu, Y. Recent Advances in Antimicrobial Polymers. Int. J. Mol. Sci. 2016, 17, 1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damodaran, V.B.; Murthy, N.S. Bio-inspired strategies for designing antifouling biomaterials. Biomater. Res. 2016, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callow, M.E.; Fletcher, R.L. The influence of low surface energy materials on bioadhesion—A review. Int. Biodeterior. Biodegrad. 1994, 34, 333–348. [Google Scholar] [CrossRef]
- Timofeeva, L.; Kleshcheva, N. Antimicrobial polymers: Mechanism of action, factors of activity, and applications. Appl. Microbiol. Biotechnol. 2011, 89, 475–492. [Google Scholar] [CrossRef]
- Liu, L.; Li, W.; Liu, Q. Recent development of antifouling polymers: Structure, evaluation, and biomedical applications in nano /micro-structures. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014, 6. [Google Scholar] [CrossRef]
- Abbasi, F.; Mirzadeh, H.; Katbab, A. Modification of polysiloxane polymers for biomedical applications: A review. Polym. Int. 2001, 1287. [Google Scholar] [CrossRef]
- Sun, W.; Liu, W.; Wu, Z.; Chen, H. Chemical Surface Modification of Polymeric Biomaterials for Biomedical Applications. Macromol. Rapid Commun. 2020, 41, 1900430. [Google Scholar] [CrossRef]
- Hons, J.C. The Development of Novel Antifouling Materials: A Multi-Disciplinary Approach. Ph.D. Thesis, Dublin City University, Dublin, Ireland, 2011. [Google Scholar]
- Lu, G.; Wu, D.; Fu, R. Studies on the synthesis and antibacterial activities of polymeric quaternary ammonium salts from dimethylaminoethyl methacrylate. React. Funct. Polym. 2007, 67, 355–366. [Google Scholar] [CrossRef]
- Elena, P.; Miri, K. Formation of contact active antimicrobial surfaces by covalent grafting of quaternary ammonium compounds. Colloids Surf. B Biointerfaces 2018, 169, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Kenawy, E.R.; Worley, S.D.; Broughton, R. The Chemistry and Applications of Antimicrobial Polymers: A State-of-the-Art Review. Biomacromolecules 2007, 8. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; de Melo Carrasco, L.D. Cationic antimicrobial polymers and their assemblies. Int. J. Mol. Sci. 2013, 14, 9906–9946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szkudlarek, M.; Heine, E.; Keul, H.; Beginn, U.; Möller, M. Synthesis, characterization, and antimicrobial properties of peptides mimicking copolymers of maleic anhydride and 4-methyl-1-pentene. Int. J. Mol. Sci. 2018, 19, 2617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, W.; Cheng, W.; Wang, G.; Liu, Y. Developments in antimicrobial polymers. J. Polym. Sci. Part A Polym. Chem. 2017, 55, 632–639. [Google Scholar] [CrossRef]
- Wu, C.; Zhou, Y.; Wang, H.; Hu, J.; Wang, X. Formation of antifouling functional coating from deposition of a zwitterionic-co-nonionic polymer via “grafting to” approach. J. Saudi Chem. Soc. 2019. [Google Scholar] [CrossRef]
- Alamri, A.; El-newehy, M.H.; Al-deyab, S.S. Biocidal polymers: Synthesis and antimicrobial properties of benzaldehyde derivatives immobilized onto amine-terminated polyacrylonitrile. Chem. Cent. J. 2012, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Bonilla, A.; López, D.; Fernández-García, M. Providing antibacterial activity to poly(2-hydroxy ethyl methacrylate) by copolymerization with a methacrylic thiazolium derivative. Int. J. Mol. Sci. 2018, 19, 4120. [Google Scholar] [CrossRef] [Green Version]
- Taylor, P.; Zhou, Z.; Calabrese, D.R.; Taylor, W.; Finlay, J.A.; Callow, M.E.; Callow, J.A.; Fischer, D.; Kramer, E.J.; Ober, C.K. Amphiphilic triblock copolymers with PEGylated hydrocarbon structures as environmentally friendly marine antifouling and fouling-release coatings. Biofouling J. Bioadhesion Biofilm 2014, 37–41. [Google Scholar] [CrossRef]
- Ward, M.; Sanchez, M.; Elasri, M.O.; Lowe, A.B. Antimicrobial activity of statistical polymethacrylic sulfopropylbetaines against gram-positive and gramnegative bacteria. J. Appl. Polym. Sci. 2006, 101, 1036–1041. [Google Scholar] [CrossRef]
- Wang, F.P.; Yuan, T.; Li, W.X.; Zhang, J.Y.; Wang, Q.Z. Synthesis and characterization of amphiphilic copolymer poly [2-(dimethylamino)ethyl methacrylate-co-methyl methacrylate]. Adv. Mater. Res. 2014, 936, 776–779. [Google Scholar] [CrossRef]
- Xu, J.; Pu, L.; Ma, J.; Kumar, S.K.; Duan, H. Antibacterial properties of synthesized cyclic and linear cationic copolymers. Polym. Chem. 2020, 11, 6632–6639. [Google Scholar] [CrossRef]
- Cuervo-Rodríguez, R.; Muñoz-Bonilla, A.; López-Fabal, F.; Fernández-García, M. Hemolytic and antimicrobial activities of a series of cationic amphiphilic copolymers comprised of same centered comonomers with thiazole moieties and polyethylene glycol derivatives. Polymers (Basel) 2020, 12, 972. [Google Scholar] [CrossRef] [PubMed]
- Martí, M.; Frígols, B.; Serrano-Aroca, A. Antimicrobial Characterization of Advanced Materials for Bioengineering Applications. JoVE 2018, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newport, S.; Differ, B.; Hoelzer, K.; Cummings, K.J.; Warnick, L.D.; Schukken, Y.H.; Siler, J.D.; Davis, M.A.; Besser, T.E.; Wiedmann, M. Agar Disk Diffusion and Automated Microbroth Dilution Produce Similar Antimicrobial Susceptibility Testing Results. Foodborne Pathog. Dis. 2011, 8. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Logan, B.E. Bacterial adhesion to glass and metal-oxide surfaces. Colloids Surf. B Biointerfaces 2004, 36, 81–90. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, P.; Chen, Z.; Cai, S.; Jing, T.; Fan, H.; Mo, F.; Zhang, J.; Lin, R. Preparation, characterization, and evaluation of amphotericin B-loaded MPEG-PCL-g-PEI micelles for local treatment of oral candida albicans. Int. J. Nanomed. 2017, 12, 4269–4283. [Google Scholar] [CrossRef] [Green Version]
- Mai, T.; Rakhmatullina, E.; Bleek, K.; Boye, S.; Yuan, J.; Völkel, A.; Gräwert, M.; Cheaib, Z.; Eick, S.; Günter, C.; et al. Poly(ethylene oxide)- b -poly(3-sulfopropyl methacrylate) block copolymers for calcium phosphate mineralization and biofilm inhibition. Biomacromolecules 2014, 15, 3901–3914. [Google Scholar] [CrossRef]
- Chen, Q.; Li, S.; Feng, Z.; Wang, M.; Cai, C.; Wang, J.; Zhang, L. Poly(2-(diethylamino)ethyl methacrylate)-based, ph-responsive, copolymeric mixed micelles for targeting anticancer drug control release. Int. J. Nanomed. 2017, 12, 6857–6870. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.; Hashmi, M.U.; Khalid, N.; Hayat, M.Q.; Ikram, A.; Janjua, H.A. Controlled assembly of silver nano-fluid in Heliotropium crispum extract: A potent anti-biofilm and bactericidal formulation. Appl. Surf. Sci. 2016, 387, 317–331. [Google Scholar] [CrossRef]
- Degirmenci, M. Synthesis and characterization of novel well-defined end-functional macrophotoinitiator of poly(MMA) by ATRP. J. Macromol. Sci. Pure Appl. Chem. 2005, 42 A, 21–30. [Google Scholar] [CrossRef]
- Zielińska, D.; Stawski, D.; Komisarczyk, A. Producing a poly(N,N-dimethylaminoethyl methacrylate) nonwoven by using the blowing out method. Text. Res. J. 2016, 86, 1837–1846. [Google Scholar] [CrossRef]
- Okten, N.S.; Canakci, C.C.; Orakdogen, N. Hertzian elasticity and triggered swelling kinetics of poly(amino ester)-based gel beads with controlled hydrophilicity and functionality: A mild and convenient synthesis via dropwise freezing into cryogenic liquid. Eur. Polym. J. 2019, 114, 176–188. [Google Scholar] [CrossRef]
- Shen Preparation and Characterization of PMMA and its Derivative via RAFT Technique in the Presence of Disulfide as a Source of Chain Transfer Agent. J. Membr. Sep. Technol. 2012, 117–128. [CrossRef]
- Huang, Y.; Yong, P.; Chen, Y.; Gao, Y.; Xu, W.; Lv, Y.; Yang, L.; Reis, R.L.; Pirraco, R.P.; Chen, J. Micellization and gelatinization in aqueous media of pH- and thermo-responsive amphiphilic ABC (PMMA82-b-PDMAEMA150- b -PNIPAM65) triblock copolymer synthesized by consecutive RAFT polymerization. RSC Adv. 2017, 7, 28711–28722. [Google Scholar] [CrossRef] [Green Version]
- Park, J.A.; Cho, K.Y.; Han, C.H.; Nam, A.; Kim, J.H.; Lee, S.H.; Choi, J.W. Quaternized Amphiphilic Block Copolymers/Graphene Oxide and a Poly(vinyl alcohol) Coating Layer on Graphene Oxide/Poly(vinylidene fluoride) Electrospun Nanofibers for Superhydrophilic and Antibacterial Properties. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.G.; Bauri, K.; Pal, S.; Goswami, A.; Madras, G.; De, P. Synthesis, characterization and thermal degradation of dual temperature- and pH-sensitive RAFT-made copolymers of N,N-(dimethylamino)ethyl methacrylate and methyl methacrylate. Polym. Int. 2013, 62, 463–473. [Google Scholar] [CrossRef]
- Georges, M.K.; Veregin, R.P.N.; Kazmaier, P.M.; Hamer, G.K. Narrow Molecular Weight Resins by a Free-Radical Polymerization Process. Macromolecules 1993, 26, 2987–2988. [Google Scholar] [CrossRef]
- Iwasaki, T.; Yoshida, J.I. Free radical polymerization in microreactors. Significant improvement in molecular weight distribution control. Macromolecules 2005, 38, 1159–1163. [Google Scholar] [CrossRef]
- Baines, F.L.; Armes, S.P.; Billingham, N.C.; Tuzar, Z. Micellization of poly(2-(dimethylamino)ethyl methacrylate-block-methyl methacrylate) copolymers in aqueous solution. Macromolecules 1996, 29, 8151–8159. [Google Scholar] [CrossRef]
- Gitchaiwat, A.; Kositchaiyong, A.; Sombatsompop, K.; Prapagdee, B.; Isarangkura, K.; Sombatsompop, N. Assessment and characterization of antifungal and antialgal performances for biocide-enhanced linear low-density polyethylene. J. Appl. Polym. Sci. 2013, 128, 371–379. [Google Scholar] [CrossRef]
- Benkhaled, B.T.; Hadiouch, S.; Olleik, H.; Perrier, J.; Ysacco, C.; Guillaneuf, Y.; Gigmes, D.; Maresca, M.; Lefay, C. Elaboration of antimicrobial polymeric materials by dispersion of well-defined amphiphilic methacrylic SG1-based copolymers. Polym. Chem. 2018, 9, 3127–3141. [Google Scholar] [CrossRef]
- Raafat, D.; Sahl, H.G. Chitosan and its antimicrobial potential—A critical literature survey. Microb. Biotechnol. 2009, 2, 186–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, P.; Dey, J.; Shome, A.; Das, P.K. Nanostructure formation in aqueous solution of amphiphilic copolymers of 2-(N,N-dimethylaminoethyl)methacrylate and alkylacrylate: Characterization, antimicrobial activity, DNA binding, and cytotoxicity studies. Int. J. Pharm. 2011, 414, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, L.A.B.; Ryan, S.M.; Mantovani, G.; Syrett, J.A.; Haddleton, D.M.; Brayden, D.J. Antibacterial effects of poly(2-(dimethylamino ethyl)methacrylate) against selected gram-positive and gram-negative bacteria. Biomacromolecules 2010, 11, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Liow, S.S.; Chee, P.L.; Owh, C.; Zhang, K.; Zhou, Y.; Gao, F.; Lakshminarayanan, R.; Loh, X.J. Cationic Poly([R]-3-hydroxybutyrate) Copolymers as Antimicrobial Agents. Macromol. Biosci. 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Oh, Y.J.; Khan, E.S.; Del Campo, A.; Hinterdorfer, P.; Li, B. Nanoscale Characteristics and Antimicrobial Properties of (SI-ATRP)-Seeded Polymer Brush Surfaces. ACS Appl. Mater. Interfaces 2019, 11, 29312–29319. [Google Scholar] [CrossRef]
- Palermo, E.F.; Sovadinova, I.; Kuroda, K. Structural determinants of antimicrobial activity and biocompatibility in membrane-disrupting methacrylamide random copolymers. Biomacromolecules 2009, 10, 3098–3107. [Google Scholar] [CrossRef]
- Takahashi, H.; Palermo, E.F.; Yasuhara, K.; Caputo, G.A.; Kuroda, K. Molecular design, structures, and activity of antimicrobial peptide-mimetic polymers. Macromol. Biosci. 2013, 13, 1285–1299. [Google Scholar] [CrossRef] [Green Version]
- Yandi, W.; Mieszkin, S.; Callow, M.E.; Callow, J.A.; Finlay, J.A.; Liedberg, B.; Ederth, T. Antialgal activity of poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA) brushes against the marine alga Ulva. Biofouling 2017, 33, 169–183. [Google Scholar] [CrossRef] [Green Version]










| Samples | Conc. (mmol) | Mol % (1HNMR) | P(DMAEMA-co-MMA) | |||
|---|---|---|---|---|---|---|
| DMAEMA | MMA | DMAEMA | MMA | Mn (g/mol) | PDI | |
| PDM 1 | 63.6 | 99.8 | 44 | 56 | 56562 | 1.75 |
| PDM 2 | 50.8 | 119.8 | 29 | 71 | 55865 | 1.65 |
| PDM 3 | 38.2 | 139.8 | 20 | 80 | 54507 | 1.93 |
| PDM 4 | 25.4 | 159.8 | 14 | 86 | 194617 | 1.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mushtaq, S.; Ahmad, N.M.; Mahmood, A.; Iqbal, M. Antibacterial Amphiphilic Copolymers of Dimethylamino Ethyl Methacrylate and Methyl Methacrylate to Control Biofilm Adhesion for Antifouling Applications. Polymers 2021, 13, 216. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13020216
Mushtaq S, Ahmad NM, Mahmood A, Iqbal M. Antibacterial Amphiphilic Copolymers of Dimethylamino Ethyl Methacrylate and Methyl Methacrylate to Control Biofilm Adhesion for Antifouling Applications. Polymers. 2021; 13(2):216. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13020216
Chicago/Turabian StyleMushtaq, Shehla, Nasir M. Ahmad, Azhar Mahmood, and Mudassir Iqbal. 2021. "Antibacterial Amphiphilic Copolymers of Dimethylamino Ethyl Methacrylate and Methyl Methacrylate to Control Biofilm Adhesion for Antifouling Applications" Polymers 13, no. 2: 216. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13020216